User:Jonathan Cardoso C. Vieira/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
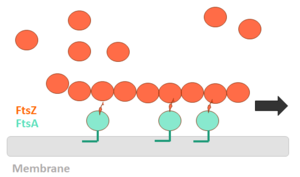
FtsZ is a tubulin-like protein, which is widely conserved in bacteria and the main component of the bacterial cytokinesis machine, or “divisome.” FtsZ is a 40 kDa protein that folds into two independent globular domains [<scene name='81/817988/N-terminal_domain/1'>N-terminal</scene> (1-203) and <scene name='81/817988/C-terminal_domain/1'>C-terminal</scene> (204-316)] and has an unstructured tail of about 50 amino acids followed by a 15–17 conserved amino acid sequence at its extreme C-terminus. This conserved terminal sequence is also known as the ‘C-terminal peptide’ (CTP), since it is in the N-terminal domain that the nucleotide binding region is contained. Self-assembly of FtsZ involves interactions between the C-terminal globular domain of one subunit with the N-terminal globular domain of another subunit. The CTP, on the other hand, is the binding site for several of the proteins that interact with FtsZ. The N-terminal and C-terminal domain are separated by the central <scene name='81/817988/H7helix/1'>H7 helix</scene> (178-202). | FtsZ is a tubulin-like protein, which is widely conserved in bacteria and the main component of the bacterial cytokinesis machine, or “divisome.” FtsZ is a 40 kDa protein that folds into two independent globular domains [<scene name='81/817988/N-terminal_domain/1'>N-terminal</scene> (1-203) and <scene name='81/817988/C-terminal_domain/1'>C-terminal</scene> (204-316)] and has an unstructured tail of about 50 amino acids followed by a 15–17 conserved amino acid sequence at its extreme C-terminus. This conserved terminal sequence is also known as the ‘C-terminal peptide’ (CTP), since it is in the N-terminal domain that the nucleotide binding region is contained. Self-assembly of FtsZ involves interactions between the C-terminal globular domain of one subunit with the N-terminal globular domain of another subunit. The CTP, on the other hand, is the binding site for several of the proteins that interact with FtsZ. The N-terminal and C-terminal domain are separated by the central <scene name='81/817988/H7helix/1'>H7 helix</scene> (178-202). | ||
FtsZ and tubulin share several essential properties: their assembly is cooperative, stimulated by GTP, and leads to GTP hydrolysis; they form dynamic polymers whose turnover is dependent on nucleotide hydrolysis; they use essentially the same bond for polymer formation; and recent evidence indicates that they undergo similar allosteric transitions upon polymerization. The folding of the FtsZ N-terminal domain is typical of GTPases, with six parallel β-strands (S1-S6) surrounded by six α-helices (H1-H6), named according to the tubulin structure (show <scene name='81/817988/Secondarystructure/2'>secondary structure</scene>). The C-terminal domain is formed by four parallel β-strands (S7-S10) surrounded by two helices, with the antiparallel strand S10. The residues in the T1-T4 loops make contact with the phosphate groups of the GDP. The T5 loop between S5 and H5 helix contains residues that make hydrogen bonds with the sugar moiety and also contacts with the phosphate of GDP, while interactions with the nucleotide nitrogen base are done by residues of the H7 helix. | FtsZ and tubulin share several essential properties: their assembly is cooperative, stimulated by GTP, and leads to GTP hydrolysis; they form dynamic polymers whose turnover is dependent on nucleotide hydrolysis; they use essentially the same bond for polymer formation; and recent evidence indicates that they undergo similar allosteric transitions upon polymerization. The folding of the FtsZ N-terminal domain is typical of GTPases, with six parallel β-strands (S1-S6) surrounded by six α-helices (H1-H6), named according to the tubulin structure (show <scene name='81/817988/Secondarystructure/2'>secondary structure</scene>). The C-terminal domain is formed by four parallel β-strands (S7-S10) surrounded by two helices, with the antiparallel strand S10. The residues in the T1-T4 loops make contact with the phosphate groups of the GDP. The T5 loop between S5 and H5 helix contains residues that make hydrogen bonds with the sugar moiety and also contacts with the phosphate of GDP, while interactions with the nucleotide nitrogen base are done by residues of the H7 helix. | ||
| - | |||
== Polymerization mechanism == | == Polymerization mechanism == | ||
Revision as of 17:38, 14 June 2019
| |||||||||||