User:Jonathan Cardoso C. Vieira/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
<StructureSection load='2vam' size='340' side='right' caption='FtsZ of Bacillus subtilis' scene=''> | <StructureSection load='2vam' size='340' side='right' caption='FtsZ of Bacillus subtilis' scene=''> | ||
== Bacillus subtilis division protein == | == Bacillus subtilis division protein == | ||
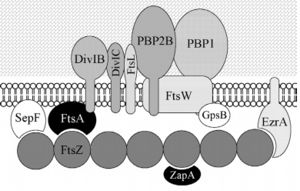
| - | Bacillus subtilis is a prokaryotic organism, a Gram-positive bacterium, and thus has a cytoplasmic membrane plus a thick cell wall made of peptidoglycan and associated anionic polymers, such as teicoic acid. Bacteria are recognized for the reproductive success and conquest of various environments on Earth, with the presence of a complex and sophisticated machinery of cell division and formation of identical daughter cells a potent factor in this success. There are currently 24 proteins known to be associated with division in B. subtilis: ClpX, DivIB,DivIC, DivIVA, EzrA, FtsA, FtsL, FtsW, FtsZ,GpsB, MciZ, MinC, MinD, MinJ, Noc, PBP1,PBP2B, SepF, SftA, SpoIIE, SpoIIIE, UgtP, YneA and ZapA. These proteins can be divided in two main groups: proteins that make up the divisome - macromolecular complex composed of about 20 proteins, which promotes the construction of the cell wall and cytoplasmic membrane, forming the division septum (DivIB, DivIC, EzrA, FtsA, FtsL, FtsW,FtsZ, GpsB, PBP1, PBP2B, SepF and ZapA) and proteins that regulate the assembly of the divisome (ClpX, DivIVA, MciZ, MinC, MinD, MinJ,Noc, UgtP and YneA). Among these, the FtsZ stands out for acting in the recruitment of other proteins for the formation of the constriction ring that culminates in cell division. | + | Bacillus subtilis is a prokaryotic organism, a Gram-positive bacterium, and thus has a cytoplasmic membrane plus a thick cell wall made of peptidoglycan and associated anionic polymers, such as teicoic acid. Bacteria are recognized for the reproductive success and conquest of various environments on Earth, with the presence of a complex and sophisticated machinery of cell division and formation of identical daughter cells a potent factor in this success. There are currently 24 proteins known to be associated with division in B. subtilis: ClpX, DivIB,DivIC, DivIVA, EzrA, FtsA, FtsL, FtsW, FtsZ,GpsB, MciZ, MinC, MinD, MinJ, Noc, PBP1,PBP2B, SepF, SftA, SpoIIE, SpoIIIE, UgtP, YneA and ZapA. These proteins can be divided in two main groups: proteins that make up the divisome - macromolecular complex composed of about 20 proteins, which promotes the construction of the cell wall and cytoplasmic membrane, forming the division septum (DivIB, DivIC, EzrA, FtsA, FtsL, FtsW,FtsZ, GpsB, PBP1, PBP2B, SepF and ZapA) and proteins that regulate the assembly of the divisome (ClpX, DivIVA, MciZ, MinC, MinD, MinJ,Noc, UgtP and YneA). |
| + | [[Image:DivisomeBacteria - Frederico.jpg | thumb | left | alt=Puzzle globe| B. subtilis divisome| 300 px]] Among these, the FtsZ stands out for acting in the recruitment of other proteins for the formation of the constriction ring that culminates in cell division. | ||
| - | == Function == | + | == General Function == |
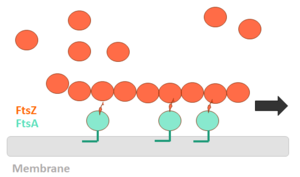
FtsZ (Filamentation temperature sensitive Z) is the main coordinator of septum formation and the most widely conserved division protein, being present in essentially all bacterial genomes that have been sequenced to date. In eubacteria, the FtsZ gene is usually found in the dcw gene cluster, a DNA region containing division and cell-wall synthesis gene. This protein is extremely important to binary fission - more specifically, formation of the Z ring - in rod-shaped bacteria entails the formation of a transverse septum that divides a progenitor cell into two equal-sized daughter cells. Septum formation is faithfully coordinated with chromosome replication and segregation and the spatial control, on the other hand, is evident in the placement of the newly formed septum at precisely the middle of the progenitor cell, which ensures that the two daughter cells generated are morphologically and genetically equivalent. It means that despite the apparent simplicity, cell division in bacteria is subject to tight spatiotemporal control. | FtsZ (Filamentation temperature sensitive Z) is the main coordinator of septum formation and the most widely conserved division protein, being present in essentially all bacterial genomes that have been sequenced to date. In eubacteria, the FtsZ gene is usually found in the dcw gene cluster, a DNA region containing division and cell-wall synthesis gene. This protein is extremely important to binary fission - more specifically, formation of the Z ring - in rod-shaped bacteria entails the formation of a transverse septum that divides a progenitor cell into two equal-sized daughter cells. Septum formation is faithfully coordinated with chromosome replication and segregation and the spatial control, on the other hand, is evident in the placement of the newly formed septum at precisely the middle of the progenitor cell, which ensures that the two daughter cells generated are morphologically and genetically equivalent. It means that despite the apparent simplicity, cell division in bacteria is subject to tight spatiotemporal control. | ||
| - | == | + | == Structure and Biochemistry of FtsZ == |
FtsZ is a tubulin-like protein, which is widely conserved in bacteria and the main component of the bacterial cytokinesis machine, or “divisome.” FtsZ is a 40 kDa protein that folds into two independent globular domains [<scene name='81/817988/N-terminal_domain/1'>N-terminal</scene> (1-203) and <scene name='81/817988/C-terminal_domain/1'>C-terminal</scene> (204-316)] and has an unstructured tail of about 50 amino acids followed by a 15–17 conserved amino acid sequence at its extreme C-terminus. This conserved terminal sequence is also known as the ‘C-terminal peptide’ (CTP), since it is in the N-terminal domain that the nucleotide binding region is contained. Self-assembly of FtsZ involves interactions between the C-terminal globular domain of one subunit with the N-terminal globular domain of another subunit. The CTP, on the other hand, is the binding site for several of the proteins that interact with FtsZ. The N-terminal and C-terminal domain are separated by the central <scene name='81/817988/H7helix/1'>H7 helix</scene> (178-202). | FtsZ is a tubulin-like protein, which is widely conserved in bacteria and the main component of the bacterial cytokinesis machine, or “divisome.” FtsZ is a 40 kDa protein that folds into two independent globular domains [<scene name='81/817988/N-terminal_domain/1'>N-terminal</scene> (1-203) and <scene name='81/817988/C-terminal_domain/1'>C-terminal</scene> (204-316)] and has an unstructured tail of about 50 amino acids followed by a 15–17 conserved amino acid sequence at its extreme C-terminus. This conserved terminal sequence is also known as the ‘C-terminal peptide’ (CTP), since it is in the N-terminal domain that the nucleotide binding region is contained. Self-assembly of FtsZ involves interactions between the C-terminal globular domain of one subunit with the N-terminal globular domain of another subunit. The CTP, on the other hand, is the binding site for several of the proteins that interact with FtsZ. The N-terminal and C-terminal domain are separated by the central <scene name='81/817988/H7helix/1'>H7 helix</scene> (178-202). | ||
FtsZ and tubulin share several essential properties: their assembly is cooperative, stimulated by GTP, and leads to GTP hydrolysis; they form dynamic polymers whose turnover is dependent on nucleotide hydrolysis; they use essentially the same bond for polymer formation; and recent evidence indicates that they undergo similar allosteric transitions upon polymerization. The folding of the FtsZ N-terminal domain is typical of GTPases, with six parallel β-strands (S1-S6) surrounded by six α-helices (H1-H6), named according to the tubulin structure (show <scene name='81/817988/Secondarystructure/2'>secondary structure</scene>). The C-terminal domain is formed by four parallel β-strands (S7-S10) surrounded by two helices, with the antiparallel strand S10. The residues in the T1-T4 loops make contact with the phosphate groups of the GDP. The T5 loop between S5 and H5 helix contains residues that make hydrogen bonds with the sugar moiety and also contacts with the phosphate of GDP, while interactions with the nucleotide nitrogen base are done by residues of the H7 helix. | FtsZ and tubulin share several essential properties: their assembly is cooperative, stimulated by GTP, and leads to GTP hydrolysis; they form dynamic polymers whose turnover is dependent on nucleotide hydrolysis; they use essentially the same bond for polymer formation; and recent evidence indicates that they undergo similar allosteric transitions upon polymerization. The folding of the FtsZ N-terminal domain is typical of GTPases, with six parallel β-strands (S1-S6) surrounded by six α-helices (H1-H6), named according to the tubulin structure (show <scene name='81/817988/Secondarystructure/2'>secondary structure</scene>). The C-terminal domain is formed by four parallel β-strands (S7-S10) surrounded by two helices, with the antiparallel strand S10. The residues in the T1-T4 loops make contact with the phosphate groups of the GDP. The T5 loop between S5 and H5 helix contains residues that make hydrogen bonds with the sugar moiety and also contacts with the phosphate of GDP, while interactions with the nucleotide nitrogen base are done by residues of the H7 helix. | ||
Revision as of 12:59, 15 June 2019
| |||||||||||
References
- ↑ Bisson-Filho AW, Discola KF, Castellen P, Blasios V, Martins A, Sforca ML, Garcia W, Zeri AC, Erickson HP, Dessen A, Gueiros-Filho FJ. FtsZ filament capping by MciZ, a developmental regulator of bacterial division. Proc Natl Acad Sci U S A. 2015 Apr 6. pii: 201414242. PMID:25848052 doi:http://dx.doi.org/10.1073/pnas.1414242112
- ↑ Wang, X. & Lutkenhaus, J. FtsZ ring: the eubacterial division apparatus conserved in archaebacteria.Mol. Microbiol. 21, 313–319 (1996). Gueiros-Filho, F. J. & Losick, R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16, 2544–2556 (2002).
- ↑ Wang, X., Huang, J., Mukherjee, A., Cao, C., and Lutkenhaus, J. (1997). Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol. 179, 5551–5559.
- ↑ Vaughan S, Wickstead B, Gull K, Addinall SG. Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J Mol Evol. 2004 Jan;58(1):19-29. doi: 10.1007/s00239-003-2523-5. PMID:14743312 doi:http://dx.doi.org/10.1007/s00239-003-2523-5
- ↑ Szwedziak P, Wang Q, Bharat TA, Tsim M, Lowe J. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. Elife. 2014 Dec 9;3:e04601. doi: 10.7554/eLife.04601. PMID:25490152 doi:http://dx.doi.org/10.7554/eLife.04601
- ↑ Huecas, S. et al. Energetics and geometry of FtsZ polymers: nucleated self-assembly of single protofilaments. Biophys. J. 94, 1796–1806 (2008).
- ↑ Lan, G. et al. (2009) Condensation of FtsZ filaments can drive bacterial cell division. Proc. Natl. Acad. Sci. U. S. A. 106, 121–126