We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
JMS/sandbox22
From Proteopedia
(Difference between revisions)
| Line 17: | Line 17: | ||
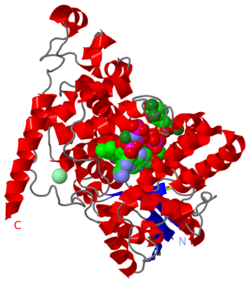
The <scene name='58/585079/1u3d_magnet/19'>cryptochrome protein</scene> absorbs a single photon of blue light of 2.7 eV, exciting either of the FAD ligand's two nitrogen atoms, which are involved in resonance (and shown in halos, as are the <scene name='58/585079/1u3d_magnet/24'>all the relevant atoms</scene>). This FAD nitrogen atom is protonated by a nearby aspartic amino acid (the proximate donors shown with halos), and the electron hole is filled through a series of electron transfers from the three tryptophan amino acids (the nitrogen donors shown with halos). Notably, as seen in this alternative view, FAD and the three tryptophans <scene name='58/585079/1u3d_magnet/21'>form a chain</scene> from the protein's inside to its outside. At this stage, where FAD is in its active signaling state, the extra electron on FAD and lone electron on the final tryptophan amino acid (324) <scene name='58/585079/1u3d_magnet/25'>have formed a radical pair</scene> (location of the electrons shown with halos). The pair is entangled, but only when they spin in the opposite directions, can the extra electron on FAD return and fill the hole left in tryptophan 324. | The <scene name='58/585079/1u3d_magnet/19'>cryptochrome protein</scene> absorbs a single photon of blue light of 2.7 eV, exciting either of the FAD ligand's two nitrogen atoms, which are involved in resonance (and shown in halos, as are the <scene name='58/585079/1u3d_magnet/24'>all the relevant atoms</scene>). This FAD nitrogen atom is protonated by a nearby aspartic amino acid (the proximate donors shown with halos), and the electron hole is filled through a series of electron transfers from the three tryptophan amino acids (the nitrogen donors shown with halos). Notably, as seen in this alternative view, FAD and the three tryptophans <scene name='58/585079/1u3d_magnet/21'>form a chain</scene> from the protein's inside to its outside. At this stage, where FAD is in its active signaling state, the extra electron on FAD and lone electron on the final tryptophan amino acid (324) <scene name='58/585079/1u3d_magnet/25'>have formed a radical pair</scene> (location of the electrons shown with halos). The pair is entangled, but only when they spin in the opposite directions, can the extra electron on FAD return and fill the hole left in tryptophan 324. | ||
| - | Researchers Klaus Schulten at University Illinois at Urbana Champaign and Ilya Solov'yov, now at the University of Southern Denmark, connect this system to the fascinating ability of many birds, and other flying species, to migrate while sensing the earth's magnetic field. Through simulations, they show that where the bird's cryptochrome compass's <scene name='58/585079/1u3d_magnet/23'>"FAD-trp324 needle"</scene> (shown as a dotted line) is aligned with the line extending between the earth's poles, the entangled electrons will 'on average' spend more time spinning the same direction, and therefore by delaying the electrons return to trp324, FAD will 'on average' be in its signalling mode for longer. | + | Researchers Klaus Schulten at University Illinois at Urbana Champaign and Ilya Solov'yov, now at the University of Southern Denmark, connect this system to the fascinating ability of many birds, and other flying species, to migrate while sensing the earth's magnetic field. Through simulations, they show that where the bird's cryptochrome compass's <scene name='58/585079/1u3d_magnet/23'>"FAD-trp324 needle"</scene> (shown as a dotted line) is aligned with the line extending between the earth's poles, the entangled electrons will 'on average' spend more time spinning in the same direction, and therefore by delaying the electrons return to trp324, FAD will 'on average' be in its signalling mode for longer. |
this is similar to what is known for <scene name='58/585079/Diamond/3'>nitrogen vacancy centers</scene>. | this is similar to what is known for <scene name='58/585079/Diamond/3'>nitrogen vacancy centers</scene>. | ||
Revision as of 20:25, 16 June 2019
| |||||||||||
References:
- ↑ Solov'yov IA, Chandler DE, Schulten K. Magnetic field effects in Arabidopsis thaliana cryptochrome-1. Biophys J. 2007 Apr 15;92(8):2711-26. Epub 2007 Jan 26. PMID:17259272 doi:http://dx.doi.org/10.1529/biophysj.106.097139
- Cryptochrome and Magnetic Sensing, Theoretical and Computational Biophysics Group at the University of Illinois at Urbana-Champaign