User:Luis Andres Casavilca Ramirez/Sandbox 1
From Proteopedia
| Line 22: | Line 22: | ||
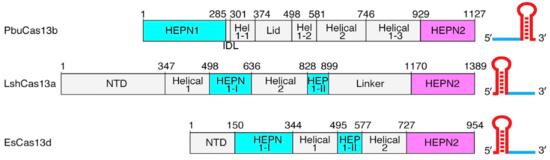
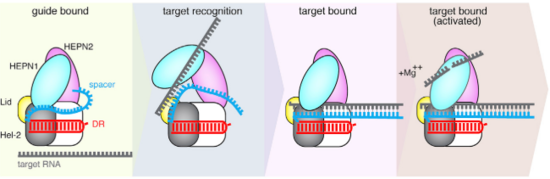
| - | The Lid domain caps the 3’ end of the direct repeat with two charged <scene name='81/817991/Lid_beta_hairpin/1'>beta-hairpins</scene>(show3’ region and two beta-hairpins of Lid domain). This region has been demonstrated to function as a second nuclease site | + | The Lid domain caps the 3’ end of the direct repeat with two charged <scene name='81/817991/Lid_beta_hairpin/1'>beta-hairpins</scene>(show3’ region and two beta-hairpins of Lid domain). This region has been demonstrated to function as a second nuclease site involved in pre-crRNA processing. Six conserved residues are predicted to coordinate and process crRNA at this second non-HEPN <scene name='81/817991/Lid_active_site/1'>catalytic site</scene>. (2)Unlike the HEPN active site, this nuclease site is thought to be metal-independent, since the activity of the equivalent crRNA processing site in Cas13a remains unaffected by the addiction of chelators and there is a formation of a 2′,3′-cyclic phosphate and a 5′-hydroxide on the 5′ and 3′ halves of the crRNA cleavage products, respectively. This last feature is characteristic of metal-independent RNA hydrolysis. (6,7) |
| - | The crRNA direct repeat consists of a <scene name='81/817991/Crrna/2'>deformed A-form duplex</scene> and is mostly buried between these two helical domains and the Lid domain, forming an extensive hydrogen bonding network which has been shown to be crucial for nuclease activity and stability of the RNA-protein complex. <scene name='81/817991/Rna_direct_repeat/1'>Highly conserved residues</scene> within Helical-2 are involved in hydrogen bonding with the nucleobases, riboses and phosphates from the direct repeat stem-loop. (*see end of paragragh) and phosphate backbone. Additionally, <scene name='81/817991/Rna_direct_repeat_a/1'>W842 also make pi-stacking interactions with U(-18)</scene> nucleobase, besides hydrogen-bonding with the phosphate backbone. <scene name='81/817991/Rna_direct_repeat/3'>Three bases</scene> | + | The crRNA direct repeat consists of a <scene name='81/817991/Crrna/2'>deformed A-form duplex</scene> and is mostly buried between these two helical domains and the Lid domain, forming an extensive hydrogen bonding network which has been shown to be crucial for nuclease activity and stability of the RNA-protein complex. <scene name='81/817991/Rna_direct_repeat/1'>Highly conserved residues</scene> within Helical-2 are involved in hydrogen bonding with the nucleobases, riboses and phosphates from the direct repeat stem-loop. (*see end of paragragh) and phosphate backbone. Additionally, <scene name='81/817991/Rna_direct_repeat_a/1'>W842 also make pi-stacking interactions with U(-18)</scene> nucleobase, besides hydrogen-bonding with the phosphate backbone. <scene name='81/817991/Rna_direct_repeat/3'>Three bases</scene> are flipped-out from the RNA body. The first two are highly important for nuclease activity and RNA-protein complex stability. C(-8) is highly conserved and important for nuclease activity and RNA-complex stability, since mutating it to G or U decreases both activity and stability of the complex. <scene name='81/817991/Rna_direct_repeat_pocket/1'>This cytosine H-bonds with T754 at the hydrophobic pocket</scene> is stabilized by interacting via its amine N4 with T754 and held in an hydrophobic pocket of hugly conserved residues (Y540, 566–571, K751, 753–761). U(-20) is absolutely conserved among Cas13b direct repeat sequences and is hydrogen-bonded to also completely conserved residues (<scene name='81/817991/Rna_direct_repeat_u20/1'>R763, R874</scene>). Mutating this nucleobase decreases both nuclease activity and thermal stability by an even greater amount than in the case of C(-8), as shown by fluorescence and thermal stability assays. The distal end of the crRNA the hairpin loop (-1 to -4 and -33 to -36) is recognized by base and backbone interactions. At least <scene name='81/817991/Rna_direct_repeat2-33-36/1'>three positions</scene> are critical for the RNase activity: U(-2), C(-36), and C(-33). U(-2) and C(-36) are contacted by N653 and N652, which coordinate the 5’ and 3’ ends of the hairpin. |
*Show the following interactions(only the ones circled in blue): | *Show the following interactions(only the ones circled in blue): | ||
| Line 48: | Line 48: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | 1. Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science (80- ). 2016;353(6299). | ||
| + | 2. Slaymaker IM, Mesa P, Kellner MJ, Kannan S, Brignole E, Koob J, et al. High-Resolution Structure of Cas13b and Biochemical Characterization of RNA Targeting and Cleavage. Cell Rep. 2019 Mar 26;26(13):3741-3751.e5. | ||
| + | 3. Wolter F, Puchta H. The CRISPR/Cas revolution reaches the RNA world: Cas13, a new Swiss Army knife for plant biologists. Plant J. 2018;94(5):767–75. | ||
| + | 4. Brennicke A. RNA editing [Internet]. Vol. 23, FEMS Microbiology Reviews. 1999. 297–316 p. Available from: http://doi.wiley.com/10.1016/S0168-6445(99)00009-1 | ||
| + | 5. Smargon AA, Cox DBT, Pyzocha NK, Zheng K, Slaymaker IM, Gootenberg JS, et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol Cell [Internet]. 2017;65(4):618-630.e7. Available from: http://dx.doi.org/10.1016/j.molcel.2016.12.023 | ||
| + | 6. East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JHD, Tjian R, et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature [Internet]. 2016;538(7624):270–3. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27669025%0Ahttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC5576363 | ||
| + | 7. Yang W. Nucleases: diversity of structure, function and mechanism. Vol. 44, Quarterly Reviews of Biophysics. 2011. 1–93 p. | ||
Revision as of 03:37, 17 June 2019
==Cas13b==
| |||||||||||
References
1. Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science (80- ). 2016;353(6299). 2. Slaymaker IM, Mesa P, Kellner MJ, Kannan S, Brignole E, Koob J, et al. High-Resolution Structure of Cas13b and Biochemical Characterization of RNA Targeting and Cleavage. Cell Rep. 2019 Mar 26;26(13):3741-3751.e5. 3. Wolter F, Puchta H. The CRISPR/Cas revolution reaches the RNA world: Cas13, a new Swiss Army knife for plant biologists. Plant J. 2018;94(5):767–75. 4. Brennicke A. RNA editing [Internet]. Vol. 23, FEMS Microbiology Reviews. 1999. 297–316 p. Available from: http://doi.wiley.com/10.1016/S0168-6445(99)00009-1 5. Smargon AA, Cox DBT, Pyzocha NK, Zheng K, Slaymaker IM, Gootenberg JS, et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol Cell [Internet]. 2017;65(4):618-630.e7. Available from: http://dx.doi.org/10.1016/j.molcel.2016.12.023 6. East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JHD, Tjian R, et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature [Internet]. 2016;538(7624):270–3. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27669025%0Ahttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC5576363 7. Yang W. Nucleases: diversity of structure, function and mechanism. Vol. 44, Quarterly Reviews of Biophysics. 2011. 1–93 p.