User:Bianca Perez Martins/Sandbox 1
From Proteopedia
| Line 29: | Line 29: | ||
==An unusual FHA/BRCT-Repeat architecture== | ==An unusual FHA/BRCT-Repeat architecture== | ||
| - | <Structure load='3huf' size='350' frame='true' align='right' caption='Structure of the S. pombe Nbs1-Ctp1 complex' | + | <Structure load='3huf' size='350' frame='true' align='right' caption='Structure of the S. pombe Nbs1-Ctp1 complex'> |
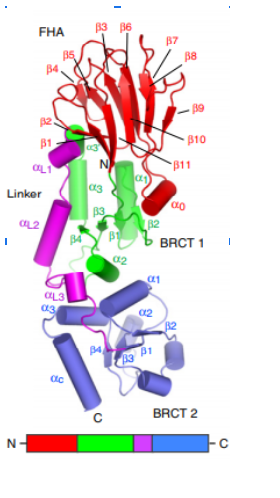
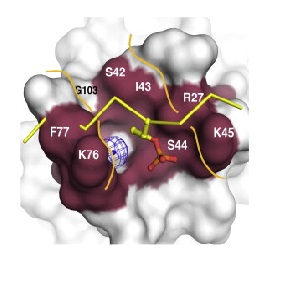
| - | One central structure for the signalling function of NBS1 is a unusual FHA/BRCT-Repeat architecture localized in the N-terminal region of this protein. This structure recognizes and binds to proteins (Mdc1 in humans, Lif1 in S. cerevisae, and | + | One central structure for the signalling function of NBS1 is a unusual FHA/BRCT-Repeat architecture localized in the N-terminal region of this protein. This structure recognizes and binds to proteins (Mdc1 in humans, Lif1 in S. cerevisae, and in S. pombe) phosphorylated in specific sites. Mutations in this domains can modify the interaction between NBS1 and these proteins, deregulating the DNA damage response. In humans, around 90% of patients suffering from NBS1 mutations has alterations on FHA/BCRT region. |
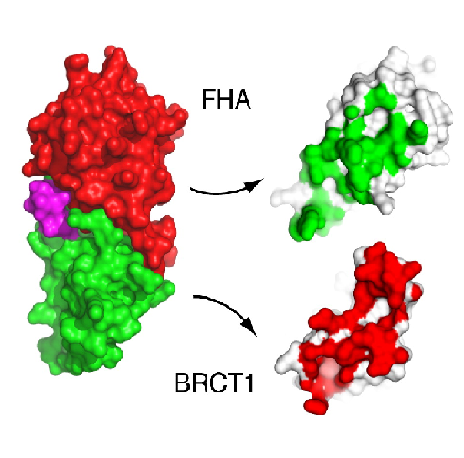
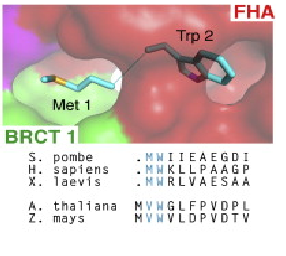
The first N-terminal 114 residues of NBS1 adopt a 𝞫 sandwitch fold characteristic of FHA domains, witch contains phospho-dependent binding modules that recognizes motifs of target proteins containing phosphothreonine (pThr). This FHA domain is suceeded by a tandem repeat of BRCT domains, phospho-protein binding domains involved in cell cycle control (Yu et al., 2003). NMR analysis of S. pombe NBS1 reveals a structural fusion of the FHA domain and first BCRT repeat, a different structure that is expected in a classical modular domains system (Lloyd et al., 2009). This association buries a substantial solvent-accessible surface of 2150 Angstrons² and its core is composed of nonpolar amino acid residues. That bulk occur between the base of FHA domain and 𝞪1, 𝞪3, and 𝞪1/𝞫2 loop of BRCT1 and stabilizes the FHA-BRCT1 as a compact structure. In this unusual case, the subsequent linker and BRCT2 domain confers more flexibility to this proteins region. | The first N-terminal 114 residues of NBS1 adopt a 𝞫 sandwitch fold characteristic of FHA domains, witch contains phospho-dependent binding modules that recognizes motifs of target proteins containing phosphothreonine (pThr). This FHA domain is suceeded by a tandem repeat of BRCT domains, phospho-protein binding domains involved in cell cycle control (Yu et al., 2003). NMR analysis of S. pombe NBS1 reveals a structural fusion of the FHA domain and first BCRT repeat, a different structure that is expected in a classical modular domains system (Lloyd et al., 2009). This association buries a substantial solvent-accessible surface of 2150 Angstrons² and its core is composed of nonpolar amino acid residues. That bulk occur between the base of FHA domain and 𝞪1, 𝞪3, and 𝞪1/𝞫2 loop of BRCT1 and stabilizes the FHA-BRCT1 as a compact structure. In this unusual case, the subsequent linker and BRCT2 domain confers more flexibility to this proteins region. | ||
| Line 44: | Line 44: | ||
===FHA/BRCT structure is highly conserved in eukaryotic NBS1=== | ===FHA/BRCT structure is highly conserved in eukaryotic NBS1=== | ||
| + | <Structure load='3hue' size='350' frame='true' align='right' caption='Structure of the S. pombe Nbs1 FHA-BRCT1-BRCT2 domains' scene='Insert optional scene name here' /> | ||
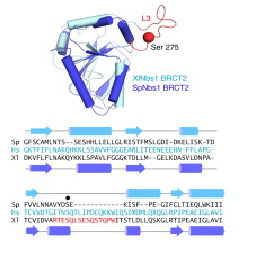
Several data indicate that this FHA/BRCT-Repeat architecture observed in spNbs1 is common to all Nbs1 orthologs, revealing the importance and conservation of its function (LlOYD et al., 2009). Comparison of eukaryotic Nbs1 N-terminal region (S. pombe, S. japonicum, S. cerevisiae, Homo sapiens and Xenopus laevis) reveals similarities in secondary and tertiary structures: 1) In all sequences the non polar character of the FHA-BRCT1 core is conserved; 2) Inside this core, one important and conserved structure is the region around the interaction of Met-1/Trp2 (buried between FHA and BRCT1 surfaces), a bulk that insulates the methionine from cotranslational excision, considering that insertions of N-terminal tags in Nbs1 generates insoluble and aggregated proteins; 3) In all this orthologs a “linker” region between the C teminus of the FHA domain and N terminus of BCRT domain is absent, indicating that this spatial apposition of FHA and BCRT domains is crucial for Nbs1 function. Comparison of Xenopus laevis, Homo sapiens and S. pombe second BRCT domain reveals remarkable similarity, but the “L3” loop which contains a major site of ATM-phosphorylation in hNbs1 (Ser-278) is replaced by a partially disordered helical loop in spNbs1 (LlOYD et al., 2009). This result can be explained by differences in this signalling pathway between these organisms. | Several data indicate that this FHA/BRCT-Repeat architecture observed in spNbs1 is common to all Nbs1 orthologs, revealing the importance and conservation of its function (LlOYD et al., 2009). Comparison of eukaryotic Nbs1 N-terminal region (S. pombe, S. japonicum, S. cerevisiae, Homo sapiens and Xenopus laevis) reveals similarities in secondary and tertiary structures: 1) In all sequences the non polar character of the FHA-BRCT1 core is conserved; 2) Inside this core, one important and conserved structure is the region around the interaction of Met-1/Trp2 (buried between FHA and BRCT1 surfaces), a bulk that insulates the methionine from cotranslational excision, considering that insertions of N-terminal tags in Nbs1 generates insoluble and aggregated proteins; 3) In all this orthologs a “linker” region between the C teminus of the FHA domain and N terminus of BCRT domain is absent, indicating that this spatial apposition of FHA and BCRT domains is crucial for Nbs1 function. Comparison of Xenopus laevis, Homo sapiens and S. pombe second BRCT domain reveals remarkable similarity, but the “L3” loop which contains a major site of ATM-phosphorylation in hNbs1 (Ser-278) is replaced by a partially disordered helical loop in spNbs1 (LlOYD et al., 2009). This result can be explained by differences in this signalling pathway between these organisms. | ||
Revision as of 03:54, 17 June 2019
Introduction: The MRN complex
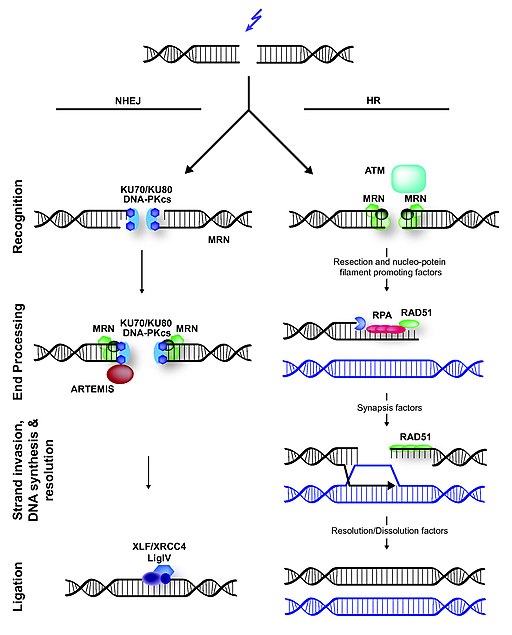
The maintenance of the DNA molecule in eukaryotes depends on several repair mechanisms that correct damages made to its structure. Important damages that can occur are Double Strand Breaks (DSBs). DSBs can be caused exposure to ionizing radiation or chemicals, or by endogenous cellular events and are some of the most significant DNA damages, because, if left unrepaired, they can result in cell death and, if misrepaired, they can cause chromosomal translocations (JEGGO, P. A.; LÖBRICH, 2007). Three processes are essential for the repair of DSBs: detection of the damage, control of the cell cycle and of transcriptional programs in response to the damage, and the presence of mechanisms for catalyzing repair of the lesion (LAMARCHE et. al. 2010). In eukaryotic cells, the MRN complex is a key factor in the response to the DSBs, since it is capable of executing the three functions mentioned above, besides being one of the earlies repair factor to bind to DSBs. There are two major pathways of DSBs repair: homologous recombination (HR) and nonhomologous end joining (NHEJ). In HR, sister chromatids are used as templates for the synthesis of the region between the ends formed by the breakage, and the use of this template makes HR a highly accurate method. However, HR can only happen during the phases of the cell cycle in which there are sister chromatids available. I contrast, in NHEJ, the DNA ends are directly ligated without the use of sister chromatids, making this repair pathway is potentially mutagenic. The MRN complex plays a role in both pathways, but interacts with different factors in each one of them. (HOPFNER, 2009).
The image illustrates the mechanism of DSBs repair in mammals. In NHEJ, broken DNA ends are rejoined, but it often requires trimming of DNA before ligation, which can lead to loss of genetic information. The trimming is thought to be done by the NMR complex. In contrast to NHEJ, HR is an error-free repair pathway that utilizes a sister chromatid, present only in the S- or G2-cell cycle phase, as template to repair DSBs. HR is initiated by DNA end-resection, involving the MRN complex and several accessory factors.
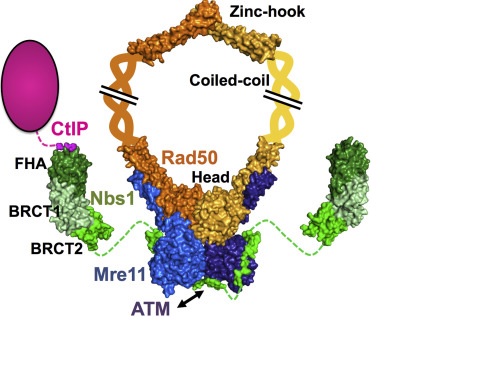
The MRN complex is formed by a dimer of Mre11, a dimer of Rad50 and a single NBS1 subunit. Mre11 acts as a nuclease, interacting directly with the DNA, Rad50 is a scaffolding component and cofactor, and NBS1 sinalizes for DNA damage response. However, Nbs1 also acts later in the repair process to regulate the DNA damage checkpoint and to recruit other repair factors to DSBs (LAFRANCE-VANASSE et. al., 2015).
The image represents the architecture of the Mre11-Rad50-Nbs1 complex with its partner CtIP. Shown in green is the Nbs1 protein, who interacts with the Mre11 dimmer (blue); The Rad50 dimer is in orange and yellow (for each monomer).
NBS1
| |||||||||||
An unusual FHA/BRCT-Repeat architecture
| |||||||||||