Introduction
PduCas13b is a class 2 type 4 from Prevotella buccae. As a member of this protein family, it consists of a single effector complexed with a CRISPR-RNA(crRNA) which binds and cleaves cleave RNA specifically, then activating a general non-specific RNase activity that degrades any nearby transcripts.(1) However, PduCas13b is structurally and mechanically different from the other Cas13s, as it has a unique linear organization and different dynamics of target binding and recognition.(2)
Though the overall conserved bilobed shape of many Class 2 effectors consisting of a recognition and nuclease lobe is present,(2,3) Cas13b has a unique domain organization even among Cas13s. (Fig.1). A crystal structure of PbuCas13b complexed with a 36-nt direct repeat sequence and a 5-nt spacer at 1.65 Å provides information of the . There are five domains within the effector structure: two HEPN domains (), two mainly helical domains (), and a . In addition, crRNA also has a unique structure, since its direct repeat is at the 3’ end and not at the 5’ end as in the other Cas13s (Fig.1).
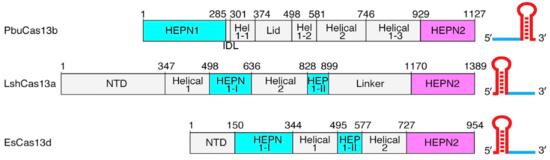
Fig.1 Linear domain organization of PbuCas13b, LshCas13a, and EsCas13d
Structure
HEPNs (Higher Eukaryotes and Prokaryotes Nucleotide-binding domain) are alpha helical domains, many of which are present in RNA maturation systems and related biological conflicts and most of which contain a catalytic Rx4-6H active site. Cas 13b domain is composed of 12 linearly connected alpha helices flexible loop between them, whereas contains nine alpha helices, several short beta strands and beta-hairpin with positively charged residues at the tip. In accordance with the conserved from HEPNs, a Rx4-6H site is present, where active residues locate within both HEPN1 () and HEPN2 () domains. One of the two histidines is thought to act as a base, inducing the ribose 2’OH to attack the phosphodiester linkage. The conserved arginine could stabilize the negative oxygens of the transition state, as a similar residue (Lysine) has been shown to carry out such function in RNase A. Alternatively, it could interact with the RNA backbone. The other polar residues, located in between the catalytic histidine and the conserved arginine, are thought to further augment the active site. (2,4) It is worth noting that, as in most CRISPR associated RNases, nuclease activity at this site is metal-dependent, as target cleavage only occurs in the presence of a certain concentration of Mg2+ and is abolished after addition of EDTA.(5) This feature contrasts with most HEPN nucleases, where activity is metal-independent. A CRISPR associated RNase, Csx2 from Pyrococcus furious, contains a Zn2+ within its HEPN domain, suggesting a possible role of a divalent cation in further stabilizing the reaction intermediate (2).
A highly conserved inter-domain linker () connects HEPN1 with Helical-1 and spans a highly positivecentral , where crRNA lies isolated from the solvent. Helical-1 domain makes extensive contacts with the direct repeat of crRNA and minor interface contacts with both HEPNs and Lid domains, whereas Helical-2, which is composed of 11 helices, makes extensive contacts with HEPN1 and minor contacts with the extended beta-hairpin of the Lid domain. Between Helical1 and Helical2 domains there is a oppositely oriented from the Lid domain. This second side channel also is positively charged and establishes access to the unbound state crRNA from the solvent.
The Lid domain caps the 3’ end of the direct repeat with two charged (show3’ region and two beta-hairpins of Lid domain). This region has been demonstrated to function as a second nuclease site involved in pre-crRNA processing. Six conserved residues are predicted to coordinate and process crRNA at this second non-HEPN . (2)Unlike the HEPN active site, this nuclease site is thought to be metal-independent, since the activity of the equivalent crRNA processing site in Cas13a remains unaffected by the addiction of chelators and there is a formation of a 2′,3′-cyclic phosphate and a 5′-hydroxide on the 5′ and 3′ halves of the crRNA cleavage products, respectively. This last feature is characteristic of metal-independent RNA hydrolysis. (6,7)
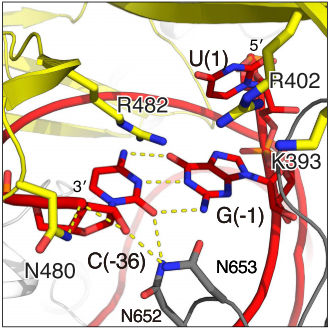
Fig. Second nuclease site
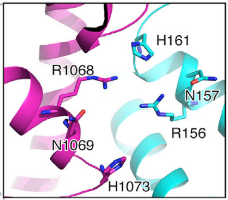
The crRNA direct repeat consists of a and is mostly buried between these two helical domains and the Lid domain, forming an extensive hydrogen bonding network which has been shown to be crucial for nuclease activity and stability of the RNA-protein complex. within Helical-2 are involved in hydrogen bonding with the nucleobases, riboses and phosphates from the direct repeat stem-loop. Additionally, nucleobase, besides hydrogen-bonding with the phosphate backbone. are flipped-out from the RNA body. The first two are highly important for nuclease activity and RNA-protein complex stability. C(-8) is highly conserved and important for nuclease activity and RNA-complex stability, since mutating it to G or U decreases both activity and stability of the complex. is stabilized by interacting via its amine N4 with T754 and held in an hydrophobic pocket of hugly conserved residues (Y540, 566–571, K751, 753–761). U(-20) is absolutely conserved among Cas13b direct repeat sequences and is hydrogen-bonded to also completely conserved residues (). Mutating this nucleobase decreases both nuclease activity and thermal stability by an even greater amount than in the case of C(-8), as shown by fluorescence and thermal stability assays. The distal end of the crRNA the hairpin loop (-1 to -4 and -33 to -36) is recognized by base and backbone interactions. At least are critical for the RNase activity: U(-2), C(-36), and C(-33). U(-2) and C(-36) are contacted by N653 and N652, which coordinate the 5’ and 3’ ends of the hairpin.
Recognition and binding dynamics of target RNA
The Cas13b-crRNA complex is practically inaccessible for hybridization with target RNA unless a major conformational shift allows access to the central channel. An opening between the domains HEPN1 and Helical-2 has been suggested, since it would provide a sterically permissible route with charged amino acids that would direct the RNA to the Cas13b central cavity. Mutation in a residue on a Lid domain β-hairpin (D397) at the interface between HEPN1 and Helical2 decreases Cas13 knockdown activity, suggesting an additional role of this domain in directing the target RNA into the central channel.
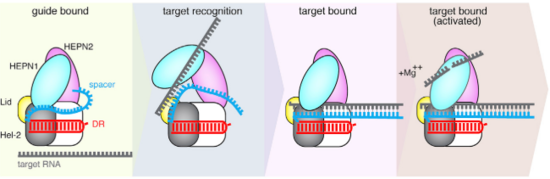
Fig.4 Proposed mechanism of crRNA targeting by Cas13b
The 3’end of target RNA is less tightly bound within the central channel and more tolerant of mismatches that the 5’end, thus being proposed as being initially recognized by the complex. In this suggested model the protein-crRNA complex initially probes the 3’RNA end of target RNA, allowing the opening of HEPN1 and Helical-2 domains and access to the central channel after complementarity is found. Then, the rest of the target RNA is hybridized. (Fig.2)
Cleavage preferentially occurs in RNA sequences with a protospacer flanking site (PFS), which consists of a non-C nucleotide at the 5’end and a AA at the 3’end, and at UU sites.(5)
Cas13b engineering
Fig.5 HEPN2 conserved loop at active site
The aforementioned preference for target RNA cleavage at dinucleotide sites is somewhat conserved among Cas 13 enzymes. A beta-hairpin loop located near the active site at HEPN2 domain is conserved among Cas13 from different species. Though the identities of its residues are variable, it is always located between highly conserved amino-acids (2) In PbuCas13b, mutants with altered residues at this loop that retained cleavage activity showed different cleavage preferences, and some of them even had increased preference for UU dinucleotides. SHERLOCK (Specific High-Sensitivity Enzymatic Reporter unLOCKing), an mRNA detection technique that takes advantage of the promiscuous cleavage activity of Cas13s is limited by the nucleobases identities of some of transcripts.(8) Therefore, Cas13b engineering at this conserved loop could potentially increase the repertoire of substrates for these type of techniques.