We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Epoxidase
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
== Structural highlights == | == Structural highlights == | ||
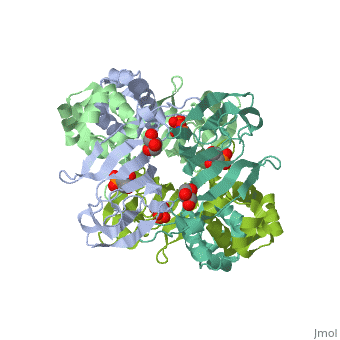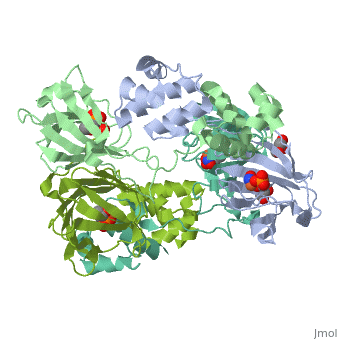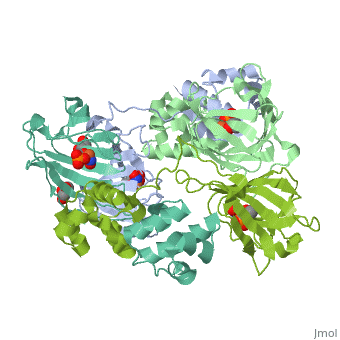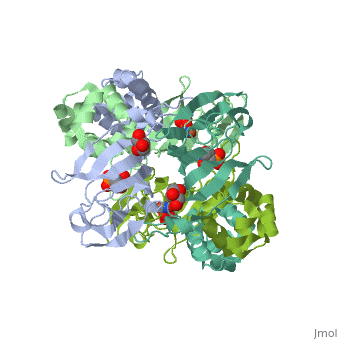
The <scene name='75/752206/Cv/6'>active site of HppE contains fosfomycin and Fe+2</scene>. The enzyme uses an induced-fit mechanism to protect high-energy iron-oxygen species formed during catalysis by moving a <scene name='75/752206/Cv/7'>β-hairpin termed cantilever</scene> to seal off the top portion of the active site<ref>PMID:21682308</ref>. <scene name='75/752206/Cv/8'>Fe coordination site</scene>. | The <scene name='75/752206/Cv/6'>active site of HppE contains fosfomycin and Fe+2</scene>. The enzyme uses an induced-fit mechanism to protect high-energy iron-oxygen species formed during catalysis by moving a <scene name='75/752206/Cv/7'>β-hairpin termed cantilever</scene> to seal off the top portion of the active site<ref>PMID:21682308</ref>. <scene name='75/752206/Cv/8'>Fe coordination site</scene>. | ||
| + | |||
| + | ==3D structures of epoxidase== | ||
| + | [[Epoxidase 3D structures]] | ||
</StructureSection> | </StructureSection> | ||
| Line 40: | Line 43: | ||
**[[3pvr]], [[3pvt]], [[3pvy]], [[3pw1]], [[3pw8]] – EcHHE subunit A + subunit C + CoA derivative <br /> | **[[3pvr]], [[3pvt]], [[3pvy]], [[3pw1]], [[3pw8]] – EcHHE subunit A + subunit C + CoA derivative <br /> | ||
**[[4ii4]] – EcHHE subunit A (mutant)+ subunit C + benzoyl CoA <br /> | **[[4ii4]] – EcHHE subunit A (mutant)+ subunit C + benzoyl CoA <br /> | ||
| + | |||
| + | * Squalene epoxidase or squalene monooxigenase | ||
| + | |||
| + | **[[6c6r]] – hSE 118-574 + FAD - human <br /> | ||
| + | **[[6c6n]], [[6c6p]] – hSE 118-574 + FAD + inhibitor <br /> | ||
* P450 epoxidase see [[Cytochrome P450]] | * P450 epoxidase see [[Cytochrome P450]] | ||
Revision as of 08:40, 23 June 2019
| |||||||||||
3D structures of epoxidase
Updated on 23-June-2019
References
- ↑ McLuskey K, Cameron S, Hammerschmidt F, Hunter WN. Structure and reactivity of hydroxypropylphosphonic acid epoxidase in fosfomycin biosynthesis by a cation- and flavin-dependent mechanism. Proc Natl Acad Sci U S A. 2005 Oct 4;102(40):14221-6. Epub 2005 Sep 26. PMID:16186494
- ↑ Watanabe F, Yu F, Ohtaki A, Yamanaka Y, Noguchi K, Yohda M, Odaka M. Crystal structures of halohydrin hydrogen-halide-lyases from Corynebacterium sp. N-1074. Proteins. 2015 Dec;83(12):2230-9. doi: 10.1002/prot.24938. Epub 2015 Oct 16. PMID:26422370 doi:http://dx.doi.org/10.1002/prot.24938
- ↑ Yun D, Dey M, Higgins LJ, Yan F, Liu HW, Drennan CL. Structural Basis of Regiospecificity of a Mononuclear Iron Enzyme in Antibiotic Fosfomycin Biosynthesis. J Am Chem Soc. 2011 Jun 30. PMID:21682308 doi:10.1021/ja2025728