We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Exonuclease
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
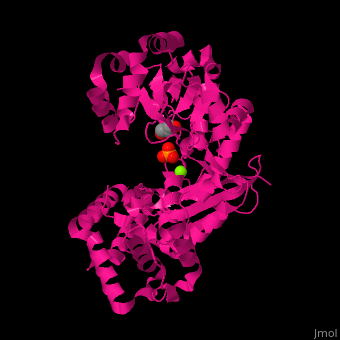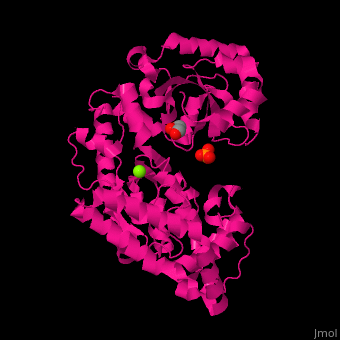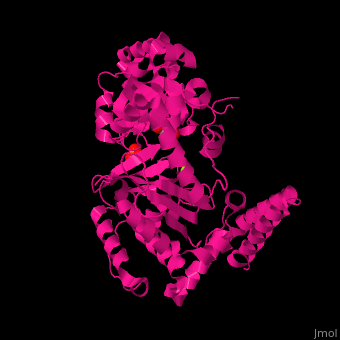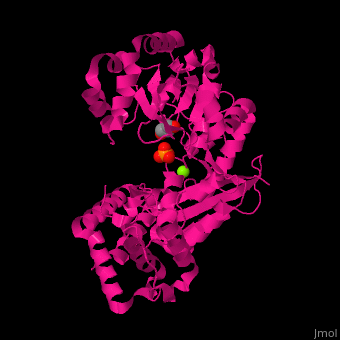
<scene name='46/466467/Cv/3'>Mg coordination site</scene> in ''E. coli'' exonuclease I (PDB code [[1fxx]]).<ref>PMID:11101894</ref> Water molecules shown as red spheres. | <scene name='46/466467/Cv/3'>Mg coordination site</scene> in ''E. coli'' exonuclease I (PDB code [[1fxx]]).<ref>PMID:11101894</ref> Water molecules shown as red spheres. | ||
| + | |||
| + | == 3D Structures of exonuclease == | ||
| + | [[Exonuclease 3D structures]] | ||
| + | |||
</StructureSection> | </StructureSection> | ||
| Line 22: | Line 26: | ||
*ExN-I | *ExN-I | ||
| + | **[[3qea]], [[3qeb]], [[5v04]], [[5v05]], [[5v06]], [[5v0b]], [[5v0c]], [[5v0d]], [[5v0e]], [[5uzv]] - hExN-I + DNA - human<br /> | ||
| + | **[[3qe9]], [[5v07]], [[5v08]], [[5v09]], [[5v0a]] – hExN-I (mutant) + DNA – human<br /> | ||
**[[1fxx]], [[3c95]] – EcExN-I – ''Escherichia coli''<br /> | **[[1fxx]], [[3c95]] – EcExN-I – ''Escherichia coli''<br /> | ||
**[[2qxf]] - EcExN-I + TMP<br /> | **[[2qxf]] - EcExN-I + TMP<br /> | ||
| Line 27: | Line 33: | ||
**[[3c94]] - EcExN-I + DNA-binding C terminal peptide<br /> | **[[3c94]] - EcExN-I + DNA-binding C terminal peptide<br /> | ||
**[[4jrp]], [[4jrq]], [[4js5]], [[4js4]], [[4hcc]], [[4hcb]] – EcExN-I + DNA<br /> | **[[4jrp]], [[4jrq]], [[4js5]], [[4js4]], [[4hcc]], [[4hcb]] – EcExN-I + DNA<br /> | ||
| - | **[[3qe9]], [[5v07]], [[5v08]], [[5v09]], [[5v0a]] – hExN-I (mutant) + DNA – human<br /> | ||
| - | **[[3qea]], [[3qeb]], [[5v04]], [[5v05]], [[5v06]], [[5v0b]], [[5v0c]], [[5v0d]], [[5v0e]], [[5uzv] - hExN-I + DNA<br /> | ||
**[[4rg8]] – ExN-I (mutant) – ''Methylocaldum szegedienes''<br /> | **[[4rg8]] – ExN-I (mutant) – ''Methylocaldum szegedienes''<br /> | ||
**[[5fiq]], [[5fis]] – ExN-I ExN domain – silkworm<br /> | **[[5fiq]], [[5fis]] – ExN-I ExN domain – silkworm<br /> | ||
| Line 89: | Line 93: | ||
*3'-5' ExN | *3'-5' ExN | ||
| + | **[[4qoz]], [[4l8r]] – hExN + histone RNA hairpin-binding protein + RNA<br /> | ||
**[[4yor]], [[4yot]], [[4you]] – PhExN phoexo I – ''Pyrococcus horikoshii''<br /> | **[[4yor]], [[4yot]], [[4you]] – PhExN phoexo I – ''Pyrococcus horikoshii''<br /> | ||
**[[4yov]], [[4yow]], [[4yox]], [[4yoy]] – PhExN phoexo I (mutant) + DNA<br /> | **[[4yov]], [[4yow]], [[4yox]], [[4yoy]] – PhExN phoexo I (mutant) + DNA<br /> | ||
| - | **[[4qoz]], [[4l8r]] – hExN + histone RNA hairpin-binding protein + RNA<br /> | ||
*5’-3’ ExN | *5’-3’ ExN | ||
| Line 104: | Line 108: | ||
**[[3mxj]] - mExN Trex1<br /> | **[[3mxj]] - mExN Trex1<br /> | ||
**[[2o4i]], [[3mxi]], [[3mxm]], [[3u3y]], [[3u6f]] - mExN Trex1 (mutant) + polynucleotide<br /> | **[[2o4i]], [[3mxi]], [[3mxm]], [[3u3y]], [[3u6f]] - mExN Trex1 (mutant) + polynucleotide<br /> | ||
| - | **[[2oa8]] - mExN Trex1 + DNA<br /> | + | **[[2oa8]], [[5ywv]], [[5ywu]], [[5ywt]], [[5yws]] - mExN Trex1 + DNA<br /> |
**[[4ynq]] - mExN Trex1 (mutant) + DNA<br /> | **[[4ynq]] - mExN Trex1 (mutant) + DNA<br /> | ||
**[[3b6o]], [[3b6p]] - mExN Trex1 + ion inhibitor<br /> | **[[3b6o]], [[3b6p]] - mExN Trex1 + ion inhibitor<br /> | ||
| + | **[[6a45]] - mExN Trex2<br /> | ||
| + | **[[6a4b]], [[6a47]] - mExN Trex2 + DNA<br /> | ||
| + | **[[6a46]] - mExN Trex2 + nucleotide<br /> | ||
*Other ExN | *Other ExN | ||
| - | **[[1ir6]], [[2zxo]], [[2zxp]], [[2zxr]] – ExN Recj – ''Thermus thermophiles''<br /> | ||
**[[1w0h]] – hExn Eri1 nuclease domain <br /> | **[[1w0h]] – hExn Eri1 nuclease domain <br /> | ||
**[[1zbu]] - hExn Eri1<br /> | **[[1zbu]] - hExn Eri1<br /> | ||
**[[1zbh]] - hExn Eri1 (mutant) + DNA<br /> | **[[1zbh]] - hExn Eri1 (mutant) + DNA<br /> | ||
**[[5aho]] – hExN Apollo<br /> | **[[5aho]] – hExN Apollo<br /> | ||
| + | **[[5zyw]] – hExN mitochondrial genome repair 1<br /> | ||
| + | **[[5zyv]], [[5zyu]], [[5zyt]] – hExN mitochondrial genome repair 1 + DNA<br /> | ||
| + | **[[3e2v]] – yExN <br /> | ||
**[[2w45]] – H4ExN alkaline - Herpesvirus 4<br /> | **[[2w45]] – H4ExN alkaline - Herpesvirus 4<br /> | ||
**[[2w4b]] - H4ExN alkaline (mutant)<br /> | **[[2w4b]] - H4ExN alkaline (mutant)<br /> | ||
| Line 120: | Line 129: | ||
**[[3sz4]] - LhExN alkaline + AMP<br /> | **[[3sz4]] - LhExN alkaline + AMP<br /> | ||
**[[3sz5]] - LhExN alkaline + polynucleotide<br /> | **[[3sz5]] - LhExN alkaline + polynucleotide<br /> | ||
| - | **[[3e2v]] – yExN <br /> | ||
**[[4lty]], [[4lu9]], [[4m0v]] – EcExN sbcD<br /> | **[[4lty]], [[4lu9]], [[4m0v]] – EcExN sbcD<br /> | ||
| + | **[[6m9k]] - ExN + recombination protein BET – Escherichia phage λ<br /> | ||
| + | **[[1ir6]], [[2zxo]], [[2zxp]], [[2zxr]] – ExN Recj – ''Thermus thermophiles''<br /> | ||
}} | }} | ||
Revision as of 07:54, 25 June 2019
| |||||||||||
3D Structures of exonuclease
Updated on 25-June-2019
References
- ↑ Mukherjee D, Fritz DT, Kilpatrick WJ, Gao M, Wilusz J. Analysis of RNA exonucleolytic activities in cellular extracts. Methods Mol Biol. 2004;257:193-212. PMID:14770007 doi:http://dx.doi.org/10.1385/1-59259-750-5:193
- ↑ Breyer WA, Matthews BW. Structure of Escherichia coli exonuclease I suggests how processivity is achieved. Nat Struct Biol. 2000 Dec;7(12):1125-8. PMID:11101894 doi:10.1038/81978