We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Gag polyprotein
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | |||
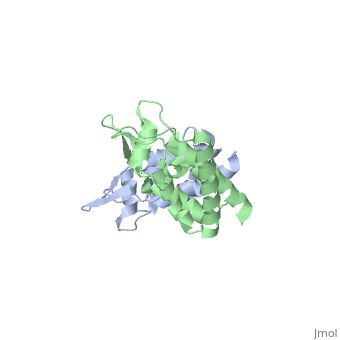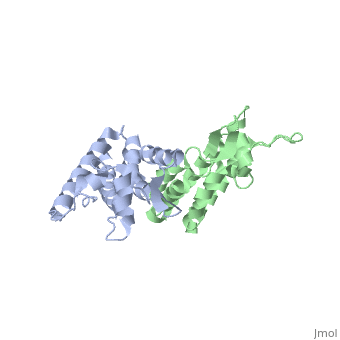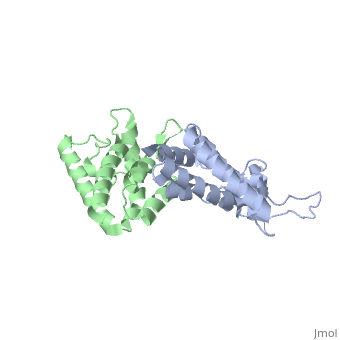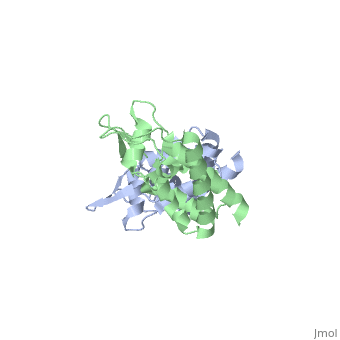
<StructureSection load='2wlv' size='350' side='right' caption='Gag polyprotein N-terminal capsid domain of HIV-2 (PDB entry [[2wlv]])' scene=''> | <StructureSection load='2wlv' size='350' side='right' caption='Gag polyprotein N-terminal capsid domain of HIV-2 (PDB entry [[2wlv]])' scene=''> | ||
| Line 17: | Line 16: | ||
==Implications== | ==Implications== | ||
HIV-1 viral particles need to form a capsid cone-like structure prior to infection of the host cell. The protealytic cleavage of the immature Gag<sup>283</sup> polyprotein results in a capsid domain. This post-translational modification is essential to the formation of the core structure. Many studies have shown that the β-hairpin formed after maturation is essential for the capsid core particle formation <ref name="gitti"/><ref name="von"/>. As a result of the β-hairpin formation, the helix 6 is displaced causing an allosteric mechanism for CpyA binding. Overall, the maturation of Gag<sup>283</sup> and formation of the mature CA protein is essential for core capsid particle creation and consequently final infection. | HIV-1 viral particles need to form a capsid cone-like structure prior to infection of the host cell. The protealytic cleavage of the immature Gag<sup>283</sup> polyprotein results in a capsid domain. This post-translational modification is essential to the formation of the core structure. Many studies have shown that the β-hairpin formed after maturation is essential for the capsid core particle formation <ref name="gitti"/><ref name="von"/>. As a result of the β-hairpin formation, the helix 6 is displaced causing an allosteric mechanism for CpyA binding. Overall, the maturation of Gag<sup>283</sup> and formation of the mature CA protein is essential for core capsid particle creation and consequently final infection. | ||
| + | |||
| + | ==3D structures of Gag polyprotein== | ||
| + | [[Gag polyprotein 3D structures]] | ||
| + | |||
</StructureSection> | </StructureSection> | ||
==3D structures of Gag polyprotein== | ==3D structures of Gag polyprotein== | ||
| Line 27: | Line 30: | ||
**[[2h3i]] – Gag residues 2-132 – HIV-1<BR /> | **[[2h3i]] – Gag residues 2-132 – HIV-1<BR /> | ||
**[[2h3f]], [[2h3i]], [[1uph]] - Gag residues 2-132 – NMR<BR /> | **[[2h3f]], [[2h3i]], [[1uph]] - Gag residues 2-132 – NMR<BR /> | ||
| + | **[[5mdg]], [[5mdf]], [[5mde]], [[5mdd]], [[5mdc]], [[5mdb]], [[5mda]], [[5md9]], [[5md8]], [[5md7]], [[5md6]], [[5md5]], [[5md4]], [[5md3]], [[5md2]], [[5md1]], [[5md0]], [[5mcz]] - Gag residues 1-221 – Cryo EM<br /> | ||
**[[1l6n]] - Gag residues 1-283 – NMR<BR /> | **[[1l6n]] - Gag residues 1-283 – NMR<BR /> | ||
**[[1gwp]] - Gag residues 132-283 – NMR<BR /> | **[[1gwp]] - Gag residues 132-283 – NMR<BR /> | ||
| Line 34: | Line 38: | ||
**[[1baj]] – Gag C terminal<BR /> | **[[1baj]] – Gag C terminal<BR /> | ||
**[[2znf]] – Gag zinc fingerlike domain - NMR<BR /> | **[[2znf]] – Gag zinc fingerlike domain - NMR<BR /> | ||
| + | **[[5teo]] - Gag residues 278-377 <br /> | ||
| + | **[[6n3j]] - Gag residues 278-377 (mutant) <br /> | ||
**[[2h3q]], [[2h3v]], [[2h3z]] - Gag residues 2-132 + phosphatidyl inositol bisphosphate - NMR<BR /> | **[[2h3q]], [[2h3v]], [[2h3z]] - Gag residues 2-132 + phosphatidyl inositol bisphosphate - NMR<BR /> | ||
**[[1mt7]], [[1mt8]] – Gag MA-CA cleavage site + protease retropepsin (mutant) <BR /> | **[[1mt7]], [[1mt8]] – Gag MA-CA cleavage site + protease retropepsin (mutant) <BR /> | ||
| Line 40: | Line 46: | ||
**[[1fgl]] – Gag residues 81-105 + cyclophilin A<BR /> | **[[1fgl]] – Gag residues 81-105 + cyclophilin A<BR /> | ||
**[[2xde]] - Gag residues 1-146 + inhibitor<BR /> | **[[2xde]] - Gag residues 1-146 + inhibitor<BR /> | ||
| - | **[[ | + | **[[4j91]], [[4j92]], [[4j93]] - Gag residues 133-278 + inhibitor<br /> |
| + | **[[6n3u]] - Gag residues 278-377 (mutant) + inhibitor<br /> | ||
**[[2x2d]] - Gag residues 133-278 + peptidyl-prolyl cis-trans isomerase A<BR /> | **[[2x2d]] - Gag residues 133-278 + peptidyl-prolyl cis-trans isomerase A<BR /> | ||
**[[2lf4]] - Gag residues 133-363 (mutant)<br /> | **[[2lf4]] - Gag residues 133-363 (mutant)<br /> | ||
| + | **[[4u0d]] - Gag residues 133-363 + Nup153 peptide<br /> | ||
| + | **[[5upw]], [[5mcy]], [[5mcx]] - Gag residues 139-351 – Cryo EM<br /> | ||
| + | **[[6ern]] - Gag residues 139-351 + ATP<br /> | ||
| + | **[[6erm]] - Gag residues 139-351 + TTP derivative<br /> | ||
| + | **[[6h09]], [[6es8]] - Gag residues 133-315 + inositol hexakisphosphate<br /> | ||
**[[2xt1]] – Gag C terminal + camelid VHH<br /> | **[[2xt1]] – Gag C terminal + camelid VHH<br /> | ||
**[[1sje]], [[1sjh]] – Gag peptide + HLA-DR1<BR /> | **[[1sje]], [[1sjh]] – Gag peptide + HLA-DR1<BR /> | ||
| Line 60: | Line 72: | ||
**[[1eoq]] - Gag C terminal – NMR<BR /> | **[[1eoq]] - Gag C terminal – NMR<BR /> | ||
**[[1a6s]] – Gag M domain (mutant) - NMR<BR /> | **[[1a6s]] – Gag M domain (mutant) - NMR<BR /> | ||
| + | **[[5a9e]] - Gag residues 84-577 – Cryo EM<br /> | ||
*Gag polyprotein from Simian immunodeficiency virus | *Gag polyprotein from Simian immunodeficiency virus | ||
| Line 69: | Line 82: | ||
**[[1u7k]], [[3bp9]] - Gag residues 215-345<BR /> | **[[1u7k]], [[3bp9]] - Gag residues 215-345<BR /> | ||
| - | **[[1u6p]] – Gag | + | **[[1u6p]] – Gag residues 479-534 + DNA <br /> |
| + | **[[6mig]] - Gag residues 683-937 + DNA<br /> | ||
*Gag polyprotein from equine infectious anemia virus | *Gag polyprotein from equine infectious anemia virus | ||
| Line 82: | Line 96: | ||
**[[4jnh]] - Gag N terminal <br /> | **[[4jnh]] - Gag N terminal <br /> | ||
| + | **[[4jmr]] - Gag N terminal + Env protein<br /> | ||
| + | **[[5m1h]], [[5m1g]] - Gag residues 300-477 - NMR<br /> | ||
}} | }} | ||
==Additional Resources== | ==Additional Resources== | ||
Revision as of 09:04, 8 July 2019
| |||||||||||
Contents |
3D structures of Gag polyprotein
Updated on 08-July-2019
Additional Resources
For additional information, see: Human Immunodeficiency Virus
Reference
- ↑ Coffin, J., S. Hughes, and H. Varmus, Retroviruses. 1997: Cold Spring Harbor Laboratory Press.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedsource - ↑ 3.0 3.1 Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996 Jul 12;273(5272):231-5. PMID:8662505
- ↑ 4.0 4.1 von Schwedler UK, Stemmler TL, Klishko VY, Li S, Albertine KH, Davis DR, Sundquist WI. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998 Mar 16;17(6):1555-68. PMID:9501077 doi:10.1093/emboj/17.6.1555
- ↑ Braaten D, Franke EK, Luban J. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J Virol. 1996 Jun;70(6):3551-60. PMID:8648689
- ↑ Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh CT, Sodroski J, Gottlinger HG. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994 Nov 24;372(6504):363-5. PMID:7969495 doi:http://dx.doi.org/10.1038/372363a0
- ↑ Ackerson B, Rey O, Canon J, Krogstad P. Cells with high cyclophilin A content support replication of human immunodeficiency virus type 1 Gag mutants with decreased ability to incorporate cyclophilin A. J Virol. 1998 Jan;72(1):303-8. PMID:9420228
Team from University of Missouri, Columbia, MO
- Students: Zheng Wang, Allison Tegge, Xin Deng
- Advisors: Jianlin Cheng, PhD, Department of Computer Science, Informatics Institute, the Life Science Center, Interdisciplinary Plant Group, University of Missouri, Columbia
- Mentor: Chun Tang, PhD, Department of Biochemistry, University of Missouri, Columbia
NMR Equipment and the Authors
Created by Allison Tegge and David Canner