Glycolate oxidase
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
{{Template:ColorKey_Turn}} | {{Template:ColorKey_Turn}} | ||
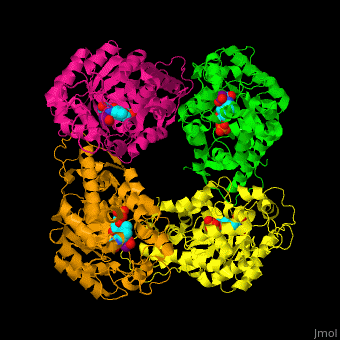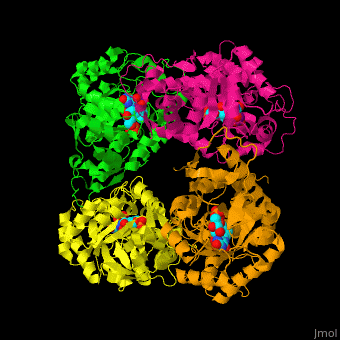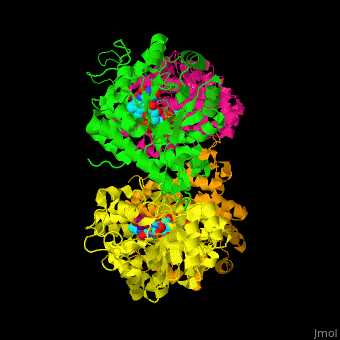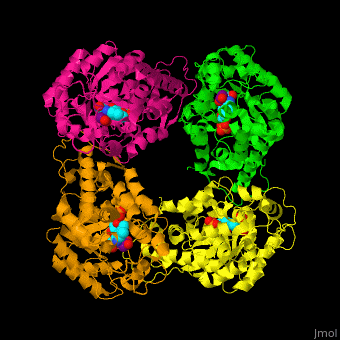
) of an α-hydroxy acid oxidase and its <scene name='48/486364/Cv/9'>active site contains the cofactor FMN</scene><ref>PMID:18215067</ref>. Water molecules are shown as red spheres. | ) of an α-hydroxy acid oxidase and its <scene name='48/486364/Cv/9'>active site contains the cofactor FMN</scene><ref>PMID:18215067</ref>. Water molecules are shown as red spheres. | ||
| + | |||
| + | ==3D structures of glycolate oxidase== | ||
| + | [[Glycolate oxidase 3D structures]] | ||
| + | |||
</StructureSection> | </StructureSection> | ||
==3D structures of glycolate oxidase== | ==3D structures of glycolate oxidase== | ||
| Line 19: | Line 23: | ||
*Glycolate oxidase | *Glycolate oxidase | ||
| - | **[[1gyl]] – sGOX – spinach<br /> | + | **[[1tb3]] – rGOX3 + FMN – rat<br /> |
| - | **[[ | + | **[[1gyl]], [[1gox]] – sGOX + FMN – spinach<br /> |
| + | **[[5zbn]] – NbGOX – ''Nicotiana benthamiana''<br /> | ||
| + | **[[5zbm]] – NbGOX + FMN <br /> | ||
**[[2yvs]] – GOX GLCE subunit – ''Thermus thermophilus''<br /> | **[[2yvs]] – GOX GLCE subunit – ''Thermus thermophilus''<br /> | ||
| - | **[[1tb3]] – rGOX3 – rat | ||
*Glycolate oxidase complex | *Glycolate oxidase complex | ||
| - | **[[ | + | **[[2rdu]], [[2nzl]] – hGOX1 + FMN + glyoxylate<br /> |
| - | **[[ | + | **[[2rdw]] – hGOX1 + FMN + sulfate<br /> |
| - | **[[ | + | **[[6gmb]] – hGOX1 + FMN + glycolate<br /> |
| - | **[[2w0u]], [[2rdt]] – hGOX1 + | + | **[[6gmc]], [[2w0u]], [[2rdt]] – hGOX1 + FMN + thiadiazole derivative<br /> |
| - | **[[3sgz]] – rGOX2 + | + | **[[5qib]], [[5qid]], [[6qif]] – hGOX1 + FMN + pyrimidine derivative<br /> |
| + | **[[5qic]] – hGOX1 + FMN + piperazine derivative<br /> | ||
| + | **[[5qie]] – hGOX1 + FMN + aniline derivative<br /> | ||
| + | **[[6qig]], [[6qih]] – hGOX1 + FMN + imidazole derivative<br /> | ||
| + | **[[3sgz]] – rGOX2 + FMN + thiadiazole derivative<br /> | ||
| + | **[[1al8]], [[1al7]] – sGOX + FMN + thiadiazole derivative<br /> | ||
**[[3giy]] – GOX1/(S)-mandelate dehydrogenase + MES – ''Pseudomonas putida'' | **[[3giy]] – GOX1/(S)-mandelate dehydrogenase + MES – ''Pseudomonas putida'' | ||
}} | }} | ||
Revision as of 09:51, 15 July 2019
| |||||||||||
3D structures of glycolate oxidase
Updated on 15-July-2019
References
- ↑ Rojas CM, Senthil-Kumar M, Wang K, Ryu CM, Kaundal A, Mysore KS. Glycolate oxidase modulates reactive oxygen species-mediated signal transduction during nonhost resistance in Nicotiana benthamiana and Arabidopsis. Plant Cell. 2012 Jan;24(1):336-52. doi: 10.1105/tpc.111.093245. Epub 2012 Jan 27. PMID:22286136 doi:http://dx.doi.org/10.1105/tpc.111.093245
- ↑ Frishberg Y, Zeharia A, Lyakhovetsky R, Bargal R, Belostotsky R. Mutations in HAO1 encoding glycolate oxidase cause isolated glycolic aciduria. J Med Genet. 2014 Aug;51(8):526-9. doi: 10.1136/jmedgenet-2014-102529. Epub 2014 , Jul 4. PMID:24996905 doi:http://dx.doi.org/10.1136/jmedgenet-2014-102529
- ↑ Murray MS, Holmes RP, Lowther WT. Active Site and Loop 4 Movements within Human Glycolate Oxidase: Implications for Substrate Specificity and Drug Design. Biochemistry. 2008 Feb 26;47(8):2439-49. Epub 2008 Jan 24. PMID:18215067 doi:10.1021/bi701710r