We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Phospholipase D
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
== Structural highlights == | == Structural highlights == | ||
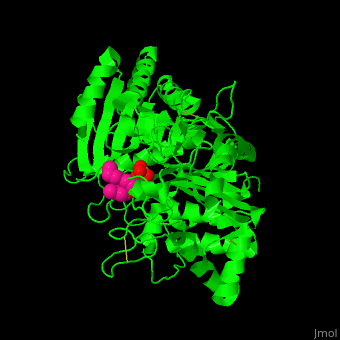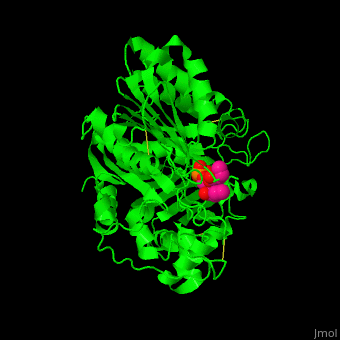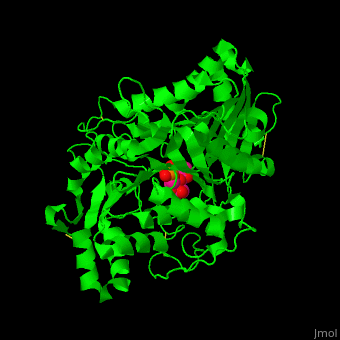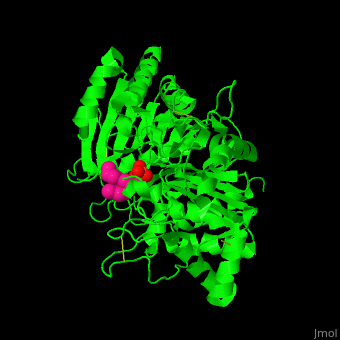
| - | The <scene name='74/749398/Cv/ | + | The <scene name='74/749398/Cv/3'>active site of PLD contains the substrate derivative dibutyrylphosphatidylcholine</scene><ref>PMID:15165852</ref>. Water molecule are shown as red sphere. |
</StructureSection> | </StructureSection> | ||
Revision as of 13:57, 4 August 2019
| |||||||||||
3D structures of phospholipase D
Updated on 04-August-2019
4rw3 – bsPLD – brown spider
4rw5 – bsPLD (mutant)
3rlg, 3rlh – PLD (mutant) – Loxosceles intemedia
4q6x – PLD – cave spider
2ze4, 1v0r, 1v0s, 1f0i – SaPLD – Streptomyces antibioticus
1v0v, 1v0w – SaPLD + phosphite
1v0y – SaPLD + phosphatidylcholine derivative
2ze9 – SaPLD (mutant) + phosphatidylcholine
1v0t, 1v0u – SaPLD + glycerophosphate
References
- ↑ McDermott M, Wakelam MJ, Morris AJ. Phospholipase D. Biochem Cell Biol. 2004 Feb;82(1):225-53. PMID:15052340 doi:http://dx.doi.org/10.1139/o03-079
- ↑ Oliveira TG, Di Paolo G. Phospholipase D in brain function and Alzheimer's disease. Biochim Biophys Acta. 2010 Aug;1801(8):799-805. doi:, 10.1016/j.bbalip.2010.04.004. Epub 2010 Apr 23. PMID:20399893 doi:http://dx.doi.org/10.1016/j.bbalip.2010.04.004
- ↑ Leiros I, McSweeney S, Hough E. The reaction mechanism of phospholipase D from Streptomyces sp. strain PMF. Snapshots along the reaction pathway reveal a pentacoordinate reaction intermediate and an unexpected final product. J Mol Biol. 2004 Jun 11;339(4):805-20. PMID:15165852 doi:http://dx.doi.org/10.1016/j.jmb.2004.04.003