We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Poly(A) RNA polymerase
From Proteopedia
(Difference between revisions)
m (Poly(A) RNA polymerase protein Cid1 moved to Poly(A) RNA polymerase: requested by Editor) |
|||
| Line 6: | Line 6: | ||
== Structural highlights == | == Structural highlights == | ||
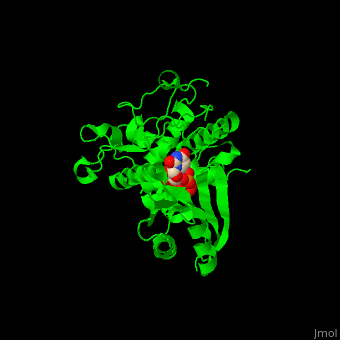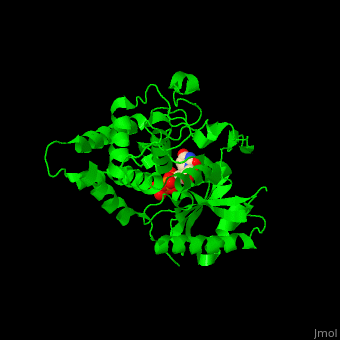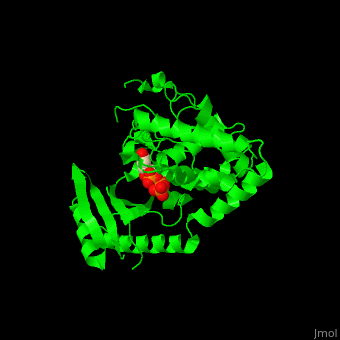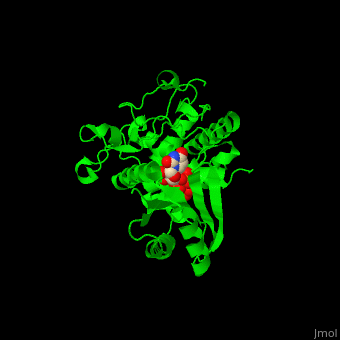
| - | The UTP binding site of Cid1 is between the N-terminal and C-terminal. The <scene name='70/706259/Cv/ | + | The UTP binding site of Cid1 is between the N-terminal and C-terminal. The <scene name='70/706259/Cv/5'>active site</scene> contains the <scene name='70/706259/Cv/6'>catalytic triad of aspartic acids</scene> (in magenta) in the N-terminal and the <scene name='70/706259/Cv/7'>His residue</scene> (in cyan) whose two conformations select between binding of UTP and ATP to Cid1 <ref>PMID:22751018</ref>. Water molecules are shown as red spheres. |
</StructureSection> | </StructureSection> | ||
Revision as of 12:52, 8 August 2019
| |||||||||||
3D Structures of poly(A) RNA polymerase protein Cid1
Updated on 08-August-2019
References
- ↑ Stevenson AL, Norbury CJ. The Cid1 family of non-canonical poly(A) polymerases. Yeast. 2006 Oct 15;23(13):991-1000. PMID:17072891 doi:http://dx.doi.org/10.1002/yea.1408
- ↑ Rouhana L, Wang L, Buter N, Kwak JE, Schiltz CA, Gonzalez T, Kelley AE, Landry CF, Wickens M. Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA. 2005 Jul;11(7):1117-30. PMID:15987818 doi:http://dx.doi.org/11/7/1117
- ↑ Yates LA, Fleurdepine S, Rissland OS, De Colibus L, Harlos K, Norbury CJ, Gilbert RJ. Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase. Nat Struct Mol Biol. 2012 Jul 1. doi: 10.1038/nsmb.2329. PMID:22751018 doi:10.1038/nsmb.2329
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky