User:Valentine J Klimkowski/Sandbox 3
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
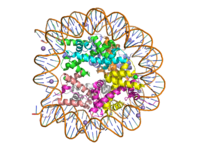
[https://en.wikipedia.org/wiki/Histone Histone proteins] aid in the packing of DNA for the purpose of compacting the genome in the nucleus of the cell and regulating physical accessibility of genes for transcription. The protein itself is an octamer of core proteins H2a, H2b, H3, and H4, which organize into two heterodimers; H1 and H5 act as linker proteins. About 145-157 base pairs wind around a histone heterodimer core. <ref name="DesJarlais">PMID: 26745824</ref> Modifications to histone core proteins can affect the accessibility of transcription factors in the genome, either promoting or inhibiting transcription. Some of these modifications include methylation/demethylation, acetylation/deacetylation, and ubiquitination/deubiquitination. <ref name="Lun">DOI: 10.1016/j.apsb.2013.04.007</ref> | [https://en.wikipedia.org/wiki/Histone Histone proteins] aid in the packing of DNA for the purpose of compacting the genome in the nucleus of the cell and regulating physical accessibility of genes for transcription. The protein itself is an octamer of core proteins H2a, H2b, H3, and H4, which organize into two heterodimers; H1 and H5 act as linker proteins. About 145-157 base pairs wind around a histone heterodimer core. <ref name="DesJarlais">PMID: 26745824</ref> Modifications to histone core proteins can affect the accessibility of transcription factors in the genome, either promoting or inhibiting transcription. Some of these modifications include methylation/demethylation, acetylation/deacetylation, and ubiquitination/deubiquitination. <ref name="Lun">DOI: 10.1016/j.apsb.2013.04.007</ref> | ||
| - | |||
| - | |||
| - | |||
Specifically, histone methylation is associated with gene activation. <ref name="Dong">PMID: 23566087</ref> Many domain families fall under the histone methylase family, one of these enzymes being the <scene name='81/811092/Set7_domain/2'>SET7 domain</scene> family, which can target H3, H4, or H2a. Sites known for gene activation are Lysine-4, Lys-36, and Lys-79 on H3; whereas, methylation at Lys-9 and Lys- 27 on H3 and Lys-20 on H4 are known for gene inactivation.<ref name="Rizzo">PMID: 21847010</ref> Typically, methylation of some of these sites are always present on both active and inactive genes, extra methylations required for activity; specifics of this characteristic depend on site and species of organism. <ref name="Xiao">doi:10.1038/nature01378</ref> Some tumor related genes such as [https://www.ncbi.nlm.nih.gov/books/NBK22268/ p53] are site specifically methylated to promote biological function <ref name = "Rizzo" />, whereas hypomethylation of [https://en.wikipedia.org/wiki/CpG_site#CpG_islands CpG] is linked to tumor genesis. <ref name="Lun" /> A particular enzyme in the SET7 domain family is lysine methyltransferase, which acts on the histone by adding a methyl group to Lys4 on H3; the addition results in promotion of gene unwinding and gene transcription. <ref name="Xiao" />, <ref name="Dong" /> | Specifically, histone methylation is associated with gene activation. <ref name="Dong">PMID: 23566087</ref> Many domain families fall under the histone methylase family, one of these enzymes being the <scene name='81/811092/Set7_domain/2'>SET7 domain</scene> family, which can target H3, H4, or H2a. Sites known for gene activation are Lysine-4, Lys-36, and Lys-79 on H3; whereas, methylation at Lys-9 and Lys- 27 on H3 and Lys-20 on H4 are known for gene inactivation.<ref name="Rizzo">PMID: 21847010</ref> Typically, methylation of some of these sites are always present on both active and inactive genes, extra methylations required for activity; specifics of this characteristic depend on site and species of organism. <ref name="Xiao">doi:10.1038/nature01378</ref> Some tumor related genes such as [https://www.ncbi.nlm.nih.gov/books/NBK22268/ p53] are site specifically methylated to promote biological function <ref name = "Rizzo" />, whereas hypomethylation of [https://en.wikipedia.org/wiki/CpG_site#CpG_islands CpG] is linked to tumor genesis. <ref name="Lun" /> A particular enzyme in the SET7 domain family is lysine methyltransferase, which acts on the histone by adding a methyl group to Lys4 on H3; the addition results in promotion of gene unwinding and gene transcription. <ref name="Xiao" />, <ref name="Dong" /> | ||
Revision as of 14:56, 12 August 2019
Histone Lysine Methyltransferase: Gene Activator
| |||||||||||
References
- ↑ DesJarlais R, Tummino PJ. Role of Histone-Modifying Enzymes and Their Complexes in Regulation of Chromatin Biology. Biochemistry. 2016 Mar 22;55(11):1584-99. doi: 10.1021/acs.biochem.5b01210. Epub , 2016 Jan 26. PMID:26745824 doi:http://dx.doi.org/10.1021/acs.biochem.5b01210
- ↑ 2.0 2.1 doi: https://dx.doi.org/10.1016/j.apsb.2013.04.007
- ↑ 3.0 3.1 Dong X, Weng Z. The correlation between histone modifications and gene expression. Epigenomics. 2013 Apr;5(2):113-6. doi: 10.2217/epi.13.13. PMID:23566087 doi:http://dx.doi.org/10.2217/epi.13.13
- ↑ 4.0 4.1 Del Rizzo PA, Trievel RC. Substrate and product specificities of SET domain methyltransferases. Epigenetics. 2011 Sep 1;6(9):1059-67. doi: 10.4161/epi.6.9.16069. Epub 2011 Sep, 1. PMID:21847010 doi:http://dx.doi.org/10.4161/epi.6.9.16069
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 5.8 Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003 Feb 6;421(6923):652-6. Epub 2003 Jan 22. PMID:12540855 doi:10.1038/nature01378
- ↑ 6.0 6.1 Kwon T, Chang JH, Kwak E, Lee CW, Joachimiak A, Kim YC, Lee J, Cho Y. Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet. EMBO J. 2003 Jan 15;22(2):292-303. PMID:12514135 doi:http://dx.doi.org/10.1093/emboj/cdg025
- ↑ 7.0 7.1 7.2 7.3 Takemoto Y, Ito A, Niwa H, Okamura M, Fujiwara T, Hirano T, Handa N, Umehara T, Sonoda T, Ogawa K, Tariq M, Nishino N, Dan S, Kagechika H, Yamori T, Yokoyama S, Yoshida M. Identification of Cyproheptadine as an Inhibitor of SET Domain Containing Lysine Methyltransferase 7/9 (Set7/9) That Regulates Estrogen-Dependent Transcription. J Med Chem. 2016 Apr 28;59(8):3650-60. doi: 10.1021/acs.jmedchem.5b01732. Epub, 2016 Apr 18. PMID:27088648 doi:http://dx.doi.org/10.1021/acs.jmedchem.5b01732
- ↑ 8.0 8.1 8.2 Tamura R, Doi S, Nakashima A, Sasaki K, Maeda K, Ueno T, Masaki T. Inhibition of the H3K4 methyltransferase SET7/9 ameliorates peritoneal fibrosis. PLoS One. 2018 May 3;13(5):e0196844. doi: 10.1371/journal.pone.0196844., eCollection 2018. PMID:29723250 doi:http://dx.doi.org/10.1371/journal.pone.0196844
Student Contributors
Lauryn Padgett,
Alexandra Pentala,
Madeleine Wilson