We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
RNase P
From Proteopedia
(Difference between revisions)
m |
|||
| Line 2: | Line 2: | ||
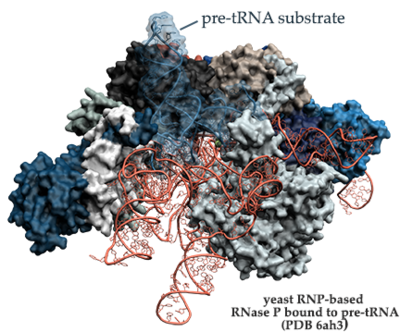
[[Image:6ah3_labeled_solid.png|left|400px]] | [[Image:6ah3_labeled_solid.png|left|400px]] | ||
== Function == | == Function == | ||
| - | '''RNase P''' processing the 5′ end of pre-[[Transfer RNA (tRNA)|transfer RNAs]] as well as other RNA molecules.<ref>PMID:28697848</ref>. Most RNase Ps are complexes of proteins and RNAs, termed ribonucleoprotein (RNP) complexes; however, a few protein-only RNase Ps have been described.<ref>PMID:23322041</ref> | + | '''RNase P''' processing the 5′ end of pre-[[Transfer RNA (tRNA)|transfer RNAs]] as well as other RNA molecules.<ref>PMID:28697848</ref>. Most RNase Ps are complexes of proteins and RNAs, termed ribonucleoprotein (RNP) complexes; however, a few protein-only RNase Ps have been described.<ref>PMID:23322041</ref><ref>PMID:25780121/ref> |
In eukaryotes, the RNase P proteins have been found to have other roles. For example, many of the proteins are shared with a related RNase P, the small nucleolar RNase MRP, that is involved in processing ribosomal RNA.<ref>PMID:19395864</ref> In yeast, the proteins of RNase P also bind telomerase.<ref>PMID:27156450</ref> | In eukaryotes, the RNase P proteins have been found to have other roles. For example, many of the proteins are shared with a related RNase P, the small nucleolar RNase MRP, that is involved in processing ribosomal RNA.<ref>PMID:19395864</ref> In yeast, the proteins of RNase P also bind telomerase.<ref>PMID:27156450</ref> | ||
| Line 20: | Line 20: | ||
Cryo-electron microscopy structure of an archaeal ribonuclease P holoenzyme. | Cryo-electron microscopy structure of an archaeal ribonuclease P holoenzyme. | ||
| - | + | ||
| + | '''RNP-based''' | ||
[[6agb]] - RNP-based RNase P - ''S. cerevisiae'' <br /> | [[6agb]] - RNP-based RNase P - ''S. cerevisiae'' <br /> | ||
[[6ah3]] - RNP-based RNase P bound to pre-tRNA substrate- ''S. cerevisiae'' <br /> | [[6ah3]] - RNP-based RNase P bound to pre-tRNA substrate- ''S. cerevisiae'' <br /> | ||
| + | |||
| + | '''Protein-based''' | ||
| + | |||
| + | Structural insights into protein-only RNase P complexed with tRNA | ||
| + | |||
Revision as of 19:55, 2 September 2019
| |||||||||||
3D Structures of RNase P
Updated on 02-September-2019
Cryo-electron microscopy structure of an archaeal ribonuclease P holoenzyme.
RNP-based
6agb - RNP-based RNase P - S. cerevisiae
6ah3 - RNP-based RNase P bound to pre-tRNA substrate- S. cerevisiae
Protein-based
Structural insights into protein-only RNase P complexed with tRNA
1jox 1jp0 1u9s
2a2e 2k3r 2ki7 2vrt
3iab 3q1q 3q1r
4g23 4g24 4g25 4g26 4xgl
4xgm
5diz 5xtm
6ahv 6bv5 6bv6 6bv8 6bv9
1a6f
1d6t
1nz0
1oqk
1ts9
1tsf
1v76
1v77
1x0t
2av5 2czv 2k3r 2ki7 2ljp 2zae
3dhs 3iab 3q1q 3q1r 3wyz 3wz0
4g23 4g24 4g25 4g26 4jg4 4xgl 4xgm
5diz 5ft9
6agb 6ah3 6ahr 6ahu 6ahv 6bv5 6bv6 6bv8 6bv9 6cqc 6cwx 6d1r 6k0a 6k0b 6max
See Also
References
- ↑ Jarrous N. Roles of RNase P and Its Subunits. Trends Genet. 2017 Sep;33(9):594-603. doi: 10.1016/j.tig.2017.06.006. Epub 2017, Jul 8. PMID:28697848 doi:http://dx.doi.org/10.1016/j.tig.2017.06.006
- ↑ Gobert A, Pinker F, Fuchsbauer O, Gutmann B, Boutin R, Roblin P, Sauter C, Giege P. Structural insights into protein-only RNase P complexed with tRNA. Nat Commun. 2013;4:1353. doi: 10.1038/ncomms2358. PMID:23322041 doi:http://dx.doi.org/10.1038/ncomms2358
- ↑ PMID:25780121/ref> In eukaryotes, the RNase P proteins have been found to have other roles. For example, many of the proteins are shared with a related RNase P, the small nucleolar RNase MRP, that is involved in processing ribosomal RNA.<ref>PMID:19395864</li> <li id="cite_note-3">[[#cite_ref-3|↑]] Lemieux B, Laterreur N, Perederina A, Noel JF, Dubois ML, Krasilnikov AS, Wellinger RJ. Active Yeast Telomerase Shares Subunits with Ribonucleoproteins RNase P and RNase MRP. Cell. 2016 May 19;165(5):1171-1181. doi: 10.1016/j.cell.2016.04.018. Epub 2016, May 5. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/27156450 27156450] doi:[http://dx.doi.org/10.1016/j.cell.2016.04.018 http://dx.doi.org/10.1016/j.cell.2016.04.018]</li></ol></ref>