RNA Polymerase II
From Proteopedia
| Line 10: | Line 10: | ||
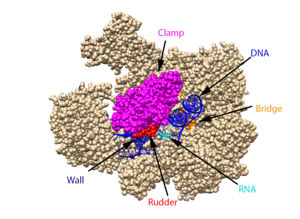
[[ Image:RnapII struct features.jpg|300px|left|thumb| The clamp (magenta), wall (navy blue), rudder (red), bridge (orange), RNA (light blue), and DNA (blue) are depicted. See below for PDB's and residue numbers.]] | [[ Image:RnapII struct features.jpg|300px|left|thumb| The clamp (magenta), wall (navy blue), rudder (red), bridge (orange), RNA (light blue), and DNA (blue) are depicted. See below for PDB's and residue numbers.]] | ||
| - | To begin, the <scene name='82/824648/Clamp/5'>clamp</scene> swings to trap the DNA in the cleft. Further along, the <scene name='82/824648/Wall/2'>wall</scene> sends the DNA template through the cleft in approximately a 90° turn. Both the clamp and wall are parts of the Rpb2 subunit. Further along in the process, the <scene name='82/824648/Rudder/ | + | To begin, the <scene name='82/824648/Clamp/5'>clamp</scene> swings to trap the DNA in the cleft. Further along, the <scene name='82/824648/Wall/2'>wall</scene> sends the DNA template through the cleft in approximately a 90° turn. Both the clamp and wall are parts of the Rpb2 subunit. Further along in the process, the <scene name='82/824648/Rudder/3'>rudder</scene> separates the newly synthesized RNA strand from the DNA template. The DNA reforms into a double helix as it leaves RNA pol II. |
Other components of RNA pol II include the following: | Other components of RNA pol II include the following: | ||
| - | The jaw is the opening through which DNA enters. The funnel is what the NTP’s travel through to be incorporated into the growing RNA strand, and the pore is the end of the funnel. The <scene name='82/824648/Bridge/4'>bridge</scene> is an Rpb1 segment that translocates the DNA-RNA complex at the end of each cycle of catalysis. <scene name='82/824648/Magnesium/ | + | The jaw is the opening through which DNA enters. The funnel is what the NTP’s travel through to be incorporated into the growing RNA strand, and the pore is the end of the funnel. The <scene name='82/824648/Bridge/4'>bridge</scene> is an Rpb1 segment that translocates the DNA-RNA complex at the end of each cycle of catalysis. <scene name='82/824648/Magnesium/3'>Magnesium</scene> is located within the active site and functions as the catalyst. |
== Mechanism of Action == | == Mechanism of Action == | ||
Revision as of 20:28, 8 October 2019
Contents |
RNAP II
| |||||||||||
References
Bushnell, D. A.; Westover, K. D.; Davis, R. E.; Kornberg, R. D. Structural Basis of Transcription: An RNA Polymerase II-TFIIB Cocrystal at 4.5 Angstroms. Science. 2004, 303, 983-988
Cramer, P.; Bushnell, D. A.; Kornberg, R. D. Structural Basis of Transcription: RNA Polymerase II at 2.8 Ångstrom Resolution. Science. 2001, 292, 1863-1876
Evans, D. A.; Fitch, D. M.; Smith, T. E.; Cee, V. J. Application of Complex Aldol Reactions to the Total Synthesis of Phorboxazole B. J. Am. Chem. Soc. 2000, 122, 10033-10046.
Gnatt, A. L.; Cramer, P; Fu, J.; Bushnell, D. A.; and Kornberg, R. D. Structural Basis of Transcription: An RNA Polymerase II Elongation Complex at 3.3 Å Resolution. Science. 2001, 292, 1876-1882 1i6h
He, Yuan, et al. Near-atomic resolution visualization of human transcription promoter opening. Nature 533.7603. 2016.
Orphanides, George, Thierry Lagrange, and Danny Reinberg. The general transcription factors of RNA polymerase II. Genes & development 10.21. 1996. 2657-2683
Uzman, A.; Voet, D. Student companion Fundamentals of biochemistry: life at the molecular level, 4th ed., Donald Voet, Judith G. Voet, Charlotte W. Pratt; John Wiley & amp; Sons, 2012.
Xu, J.; Lahiri, I.; Wang, W.; Wier, A.; Cianfrocco, M. A.; Chong, J.; Hare, A. A.; Dervan, P. B.; DiMaio, F.; Leschziner, A. E.; Wang, D. Structural Basis for the Initiation of Eukaryotic Transcription-coupled DNA Repair. Nature. 2017. 551, 653-657 5vvr
Yan, C., Dodd, T., He, Y., Tainer, J. A., Tsutakawa, S. E., & Ivanov, I. (2019). Transcription preinitiation complex structure and dynamics provide insight into genetic diseases. Nature Structural and Molecular Biology, 26(6), 397-406.
Notes
From structural components:
Structural overview: [PDB: 5VVR: with highlighted sections mentioned below]
Bridge: Depicted: [PDB: 1I6H: 810-845.a]
Wall: Depicted: [PDB: 1R5U: 853-919.b; 933-972.b]
Clamp: Depicted: [PDB: 1R5U: 3-345.a; 1395-1435.a; 1158-1124.b]
Rudder: Depicted: [PDB: 5VVR: 306-321.a]
Content Donators
This page was created as a final project for the Advanced Biochemistry course at Wabash College during the Fall of 2019. This page was reviewed by Dr. Wally Novak of Wabash College.
Proteopedia Page Contributors and Editors (what is this?)
Titus Edwards, Abraham Kiesel, Wally Novak, James Daniel Andry, Michal Harel, Karsten Theis