From Proteopedia
(Difference between revisions)
proteopedia linkproteopedia link
|
|
| Line 13: |
Line 13: |
| | | | |
| | Other components of RNA pol II include the following: | | Other components of RNA pol II include the following: |
| - | The jaw is the opening through which DNA enters. The funnel is what the NTP’s travel through to be incorporated into the growing RNA strand, and the pore is the end of the funnel. The <scene name='82/824648/Bridge/4'>bridge</scene> is an Rpb1 segment that translocates the DNA-RNA complex at the end of each cycle of catalysis. <scene name='82/824648/Magnesium/3'>Magnesium</scene> is located within the active site and functions as the catalyst. | + | The jaw is the opening through which DNA enters. The funnel is what the NTP’s travel through to be incorporated into the growing RNA strand, and the pore is the end of the funnel. The <scene name='82/824648/Bridge/6'>bridge</scene> is an Rpb1 segment that translocates the DNA-RNA complex at the end of each cycle of catalysis. <scene name='82/824648/Magnesium/3'>Magnesium</scene> is located within the active site and functions as the catalyst. |
| | | | |
| | == Mechanism of Action == | | == Mechanism of Action == |
| | | | |
| - | 1wcm <scene name='82/824544/Active_site_1/1'>TextToBeDisplayed</scene> | + | 1wcm <scene name='82/824544/Active_site_1/1'>Daniel Andry is a stud</scene> |
| - | 1i6h <scene name='82/824648/Active_site_2/1'>TextToBeDisplayed</scene> | + | 1i6h <scene name='82/824648/Active_site_2/1'>James Daniel Andry is my hero</scene> |
| | | | |
| | == α-Amanitin == | | == α-Amanitin == |
Revision as of 20:32, 8 October 2019
RNAP II
|
Introduction
RNA polymerase II (RNAP II) is an enzyme that transcribes protein-encoding genes, and it therefore is responsible for the synthesis of mRNA.Brief description of mRNA
Structural Components
This section will briefly discuss the chief structural components involved in the mechanism.
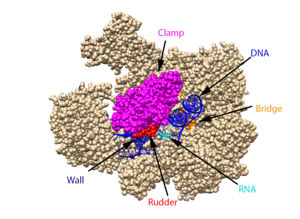 The clamp (magenta), wall (navy blue), rudder (red), bridge (orange), RNA (light blue), and DNA (blue) are depicted. See below for PDB's and residue numbers. To begin, the swings to trap the DNA in the cleft. Further along, the sends the DNA template through the cleft in approximately a 90° turn. Both the clamp and wall are parts of the Rpb2 subunit. Further along in the process, the separates the newly synthesized RNA strand from the DNA template. The DNA reforms into a double helix as it leaves RNA pol II.
Other components of RNA pol II include the following:
The jaw is the opening through which DNA enters. The funnel is what the NTP’s travel through to be incorporated into the growing RNA strand, and the pore is the end of the funnel. The is an Rpb1 segment that translocates the DNA-RNA complex at the end of each cycle of catalysis. is located within the active site and functions as the catalyst.
Mechanism of Action
1wcm
1i6h
α-Amanitin
α-Amanitin is a bicyclic octapeptide that adheres tightly with RNAP II, which blocks the elongation steps. α-amanitin binds in the funnel and interacts with the bridge helix and adjacent Rpb1, but it does not inhibit the RNA pol II’s interaction with NTP. Instead, α-amanitin likely challenges the bridge’s conformational change that is necessary for the purposed RNAP translocation step. α-Amanitin, found in the poisonous mushroom death cap, leads to death after several days. This time frame aligns with the rate at which mRNA’s and proteins turnover.
Modifications
General Transcription Factors
In both eukaryotes and prokaryotes, the basic mechanism for initiating transcription is the same: protein factors selectively bind to promoter regions on DNA. Prokaryotes use sigma factors while eukaryotes use a complex of 6 general transcription factors (GTFs). These GTFs are all named similarly and begin with TF, for transcription factor, followed by the Roman numeral II since they are involved in transcription by RNAP II. The combination of all the transcription factors bound to the DNA promoter region, in complex with RNAP II, is called the preinitiation complex (PIC). The formation of the PIC occurs in an ordered pathway, beginning with the TATA box which is a promoter region on DNA at position -27.
Process of PIC formation:
1. contains a subunit named the TATA-binding protein (TBP), which recognizes and binds to the TATA box on the DNA promoter.
2. and and interact with TBP and are recruted to the promoter.
3. binds directly to RNAP II and escorts it to the promoter while helps the complex bind correctly.
4. and are sequentually recruited which completes the PIC.
PIC NON-TRANSPARENT COMPLEX
FIGURE CAPTIONS-PDB AND COLOR
CENTERING F
Once the PIC is formed, RNAP II initiates RNA synthesis and produces a short transcript. When RNAP II becomes phosphorylated, it releases some of the GTFs from the complex and moves away from the promoter. TFIID stays bound to the promoter and can reinitiate transcription. The transcription factors are replaced by a new six-protein complex call the Elongator. TFIIF and TFIIH both remain associated with RNAP II during elongation.
|
References
Bushnell, D. A.; Westover, K. D.; Davis, R. E.; Kornberg, R. D. Structural Basis of Transcription: An RNA Polymerase II-TFIIB Cocrystal at 4.5 Angstroms. Science. 2004, 303, 983-988
Cramer, P.; Bushnell, D. A.; Kornberg, R. D. Structural Basis of Transcription: RNA Polymerase II at 2.8 Ångstrom Resolution. Science. 2001, 292, 1863-1876
Evans, D. A.; Fitch, D. M.; Smith, T. E.; Cee, V. J. Application of Complex Aldol Reactions to the Total Synthesis of Phorboxazole B. J. Am. Chem. Soc. 2000, 122, 10033-10046.
Gnatt, A. L.; Cramer, P; Fu, J.; Bushnell, D. A.; and Kornberg, R. D. Structural Basis of Transcription: An RNA Polymerase II Elongation Complex at 3.3 Å Resolution. Science. 2001, 292, 1876-1882 1i6h
He, Yuan, et al. Near-atomic resolution visualization of human transcription promoter opening. Nature 533.7603. 2016.
Orphanides, George, Thierry Lagrange, and Danny Reinberg. The general transcription factors of RNA polymerase II. Genes & development 10.21. 1996. 2657-2683
Uzman, A.; Voet, D. Student companion Fundamentals of biochemistry: life at the molecular level, 4th ed., Donald Voet, Judith G. Voet, Charlotte W. Pratt; John Wiley & amp; Sons, 2012.
Xu, J.; Lahiri, I.; Wang, W.; Wier, A.; Cianfrocco, M. A.; Chong, J.; Hare, A. A.; Dervan, P. B.; DiMaio, F.; Leschziner, A. E.; Wang, D. Structural Basis for the Initiation of Eukaryotic Transcription-coupled DNA Repair. Nature. 2017. 551, 653-657 5vvr
Yan, C., Dodd, T., He, Y., Tainer, J. A., Tsutakawa, S. E., & Ivanov, I. (2019). Transcription preinitiation complex structure and dynamics provide insight into genetic diseases. Nature Structural and Molecular Biology, 26(6), 397-406.
Notes
From structural components:
Structural overview: [PDB: 5VVR: with highlighted sections mentioned below]
Bridge: Depicted: [PDB: 1I6H: 810-845.a]
Wall: Depicted: [PDB: 1R5U: 853-919.b; 933-972.b]
Clamp: Depicted: [PDB: 1R5U: 3-345.a; 1395-1435.a; 1158-1124.b]
Rudder: Depicted: [PDB: 5VVR: 306-321.a]
Content Donators
This page was created as a final project for the Advanced Biochemistry course at Wabash College during the Fall of 2019. This page was reviewed by Dr. Wally Novak of Wabash College.

