Uracil glycosylase inhibitor
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
== Structural highlights == | == Structural highlights == | ||
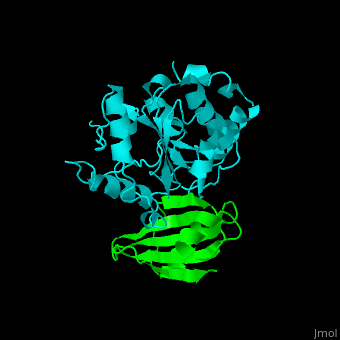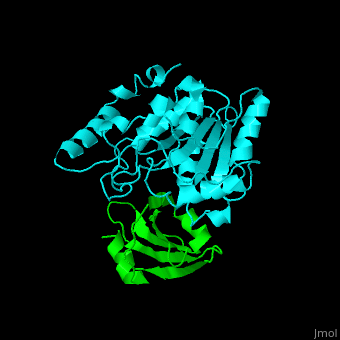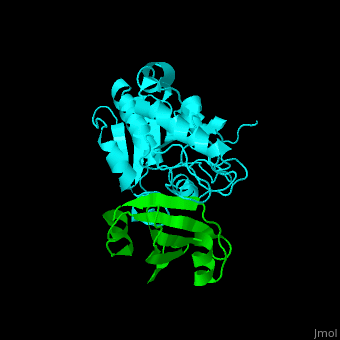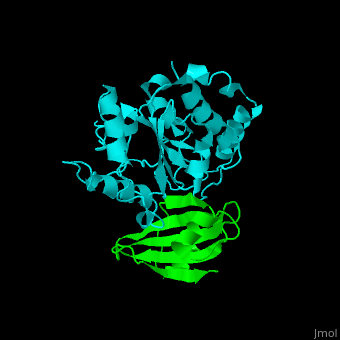
| - | The <scene name='51/516470/Cv/ | + | The <scene name='51/516470/Cv/3'>surface interactions between the UDG and UGI is fairly polar</scene><ref>PMID:10080896</ref> ({{Template:ColorKey_Hydrophobic}}, {{Template:ColorKey_Polar}}). Interacting UGI residues shown in ball-and-stick representation, UDG residues shown in spacefill representation. |
==3D structures of uracil glycosylase inhibitor== | ==3D structures of uracil glycosylase inhibitor== | ||
Revision as of 12:08, 10 October 2019
| |||||||||||
References
- ↑ Acharya N, Kumar P, Varshney U. Complexes of the uracil-DNA glycosylase inhibitor protein, Ugi, with Mycobacterium smegmatis and Mycobacterium tuberculosis uracil-DNA glycosylases. Microbiology. 2003 Jul;149(Pt 7):1647-58. PMID:12855717 doi:http://dx.doi.org/10.1099/mic.0.26228-0
- ↑ Putnam CD, Shroyer MJ, Lundquist AJ, Mol CD, Arvai AS, Mosbaugh DW, Tainer JA. Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase. J Mol Biol. 1999 Mar 26;287(2):331-46. PMID:10080896 doi:10.1006/jmbi.1999.2605