Cytochromes
Cytochrome b
Cytochrome b5
Cytochrome b5 (CB) functions as an electron transport carrier for several membrane-bound oxygenases. CB is heme-containing protein. The microsomal and mitochondrial CB are membrane-bound while bacterial and other animal tissue CB are soluble. Cytochrome b562 is the the b-type cytochrome from E. coli.[1] (PDB entry 1b5m[2]) is shown.
Cytochrome bc1 complex
Cytochrome bc1 (Cbc1) functions as the central pump which transfers protons across the cell membrane. The protons are used to power the rotation of ATP synthase. Cbc1 binds ubiquinol which carries hydrogen atoms. Cbc1 separates the protons and the electrons. The protons are released in the inner side of the membrane for use by ATP synthase and the electrons are transferred to cytochrome c or to the outer side of the membrane. Plants use cytochrome b6f in the same manner binding plastoquinol as a hydrogen carrier. Stigmatellin inhibits the Cbc1 electron transfer by binding to its quinone oxidation site. Antimycin inhibits Cbc1 by binding to its quinone reduction site.[3]
More details in Complex_III_of_Electron_Transport_Chain.
Structural highlights
Cbc1 is a composed of 11 proteins and cofactors which include heme-carrying proteins like and and iron-sulfur cluster proteins like . The iron containing moieties are , (where vinyl side chain of heme are replaced by thioether) and . [4]
Cytochrome c
Structural and kinetic studies of imidazole binding to two members of the cytochrome c6 family reveal an important role for a conserved heme pocket residue[5]
is a member of the class I family of c-type cytochromes with a distinctive and a . They function in the photosynthetic electron transport chain of cyanobacteria where they shuttle an electron from the cytochrome b6f complex to photosystem I. Structures of numerous cytochrome c6 proteins have been determined and all have the . In the present work we have solved the structure of the Q51V site-directed variant of Phormidium laminosum cytochrome c6. This project is part of a study that is aimed at gaining insight into protein factors which modulate the heme mid-point redox potential in the cytochrome c6 family. The Q51V variant has been shown to tune over 100 mV of heme redox potential, which for a single heme pocket mutation is very significant and has consequences for function.
The Q51V structure confirms that the has the same side-chain orientation in the heme pocket as found in other cytochrome c6 proteins, that naturally have a Val at this position. The significance of this structure is that the and an . Two other structures of imidazole cyt c-adducts have been reported, but neither appear to undergo the . Both protein and heme structural changes are observed, with the later centered on a accompanied by the and the .
Protein (un)folding studies on cytochrome c have revealed that (un)folding involves structural units called 'foldons'. The regions in the Q51V imidazole-adduct where structural changes occur map well to the two foldons predicted to unfold first in cytochrome c. Thus , leading to the formation of an early unfolding intermediate that is stabilised by , enabling it to be captured in the crystalline form.
Structural model of the [Fe]-hydrogenase/cytochrome C553 complex combining NMR and soft-docking[6]
The shows the specific interaction of the hydrogenase (light blue) with the cytochrome (pink), revealing the path of electron transport from the , through three iron-sulfur clusters, and ending in the cytochrome heme (colored red). Two , CYS 38 in the hydrogenase and CYS10 in the cytochrome, are thought to provide the electron transfer pathway between the two proteins (these scenes were created by Jaime Prilusky, David S. Goodsell, and Eran Hodis).
Conformational control of the binding of diatomic gases to cytochrome c’ [7]
The cytochromes c′ (CYTcp) are found in denitrifying, methanotrophic and photosynthetic bacteria. These proteins are able to form stable adducts with CO and NO but not with O2. The binding of NO to CYTcp currently provides the best structural model for the NO activation mechanism of soluble guanylate cyclase. Ligand binding in CYTcps has been shown to be highly dependent on residues in both the proximal and distal heme pockets. Group 1 CYTcps typically have a phenylalanine residue positioned close to the distal face of heme, while for group 2, this residue is typically leucine. We have structurally, spectroscopically and kinetically characterised the CYTcp from Shewanella frigidimarina , a protein that has a distal phenylalanine residue and a lysine in the proximal pocket in place of the more common arginine (monomer A is colored in red, monomer B in green, and heme group in yellow). in a similar manner to CYTcps previously characterised.
SFCP exhibits biphasic binding kinetics for both NO and CO as a result of the high level of steric hindrance from the aromatic side chain of residue Phe 16. The binding of distal ligands is thus controlled by the conformation of the phenylalanine ring.
- of SFCP (in green;4ulv), RCCP (R. capsulatus; in magenta; 1cpq), RSCP (R. sphaeroides; in red; 1gqa) and RGCP (R. gelatinosus; in cya); 2j8w).
- .
Only a proximal 5-coordinate NO adduct, confirmed by structural data, is observed with no detectable hexacoordinate distal NO adduct.
Rhodothermus marinus cytochrome c
Structure
All members in the C-type cytochrome superfamily contain a heme prosthetic group that is covalently attached to the protein via two thioether bonds to cysteine residues. Most cytochromes c occur in a where the histidine residue is one of the two axial ligands of the heme iron.[8][9] In monoheme cytochromes c, the other axial position may be left vacant or be occupied by histidine or methionine residues; however, it can sometimes be occupied by cysteine or lysine residues.[8]. In Rmcytc, XX represents a threonine (Thr46) and an alanine residue (Ala47) that help form the loop 2 structure.
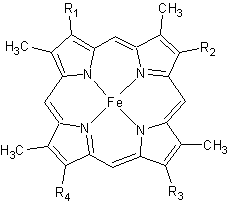
Figure 2. The tetrapyrrolic heme prosthetic group that can either be covalently attached to or closely associated with various proteins, such as cytochromes and other globin proteins. In Rmcytc, R2 is an ethyl group covalently attached to Cys 45, and R3 is a methyl group covalently attached to Cys48.
The typical monoheme cyt c fold is formed by helices . Rmcytc contains seven α-helices that are folded around the heme, all connected by random coils.[8] The heme group is axially coordinated by , and the disulfide linkages exist at . The heme group in Rmcytc is almost completely shielded from solvent due to it being in a mostly hydrophobic pocket. This pocket is formed in part by the seven helices surrounding the ring, but also by two structures that are uncommon in other cytochromes c. First, a 21 amino acid extension of the N-terminal exists, forming , which wraps around the back of the polypeptide.[8] An extension resembling such has only been seen in Thermus thermophilus; however, the extension occurs at the C-terminus rather than the N-terminus.[10] A second rarity is that of , inserted between helix D and loop 3, that shields the bottom part of the heme from any solvent.[8] In cytochrome c2 as well as mitochondrial cyt c, a similar yet shorter helix was found, though this helix was present at a different place in the primary sequence. Also, instead of helix B', T. thermophilus contains a two-stranded β-sheet.[8] One final note is the number of residues that Rmcytc contains. In general, cyt c contains about two methionines whereas Rmcytc contains seven, located on the left of the heme.[8]
As determined by X-ray crystallography, the Rmcytc structure was found to contain a sulfate ion coordinated to Glu122 via hydrogen bonding to the protonated carboxylate oxygen. In the protein complex, this ion has been seen to mediate crystal contact between neighbouring protein molecules.[8]
The observation of these structural motifs in other C-type cytochromes can support the divergent evolution of cytochromes c.[8] These motifs are present in a number of different bacteria and are seen in similar regions of the secondary structure; however, they exist in the primary sequence in places distinct to the phylum. For example, monoheme cytochromes c in the rest of the Bacteroidetes phylum have an N-terminus extension that is highly conserved to that of Rmcytc, and the regions in the primary structure that correspond to these secondary motifs are not observed in other bacterial phyla.[8] Also, due to these motifs being absent from other phyla, the Bacteroidetes monoheme cyt c has been said to form a new subfamily of cyt c.
Function
Monoheme cytochromes c are involved in electron transport chains in both prokaryotes and eukaryotic mitochondria.[8] They mediate the transfer of electrons mainly from the bc1 complexes or their analogs to heme-copper oxygen reductases (HCOs) in the electron transport chain of oxidative phosphorylation. Heme c containing domains are often found fused to other protein domains such as these HCOs, including the caa3 oxygen reductases[8][11]; these enzymes are membrane-bound and catalyze the reduction of O2 to water.[12] In addition to being involved in oxidative phosphorylation, monoheme cyt c has also been seen to participate in the electron transport chain of photosynthesis.[8] Cytochrome c has also been determined to be a major signalling molecule in the apoptotic pathways.
Electron transport chain
In the electron transport chain (ETC), cyt c shuttles electrons between the respiratory complexes III and IV; complex III is the cytochrome bc1 complex and IV is cyt c oxidase. Initially, the heme iron in cyt c is in the reduced, Fe3+ state; this allows for the uptake of one electron, oxidizing the iron to the Fe2+ state.[13] The ETC in eukaryotes is quite simple compared to that of prokaryotes (Figure 3).
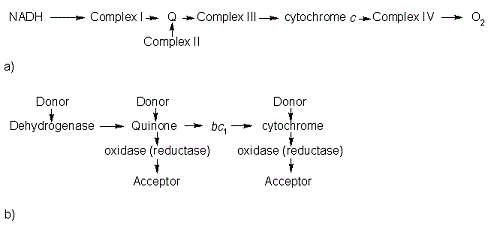
Figure 3. The electron transport chain of a) eukaryotes as compared to b) prokaryotes.
In prokaryotic systems, electrons can enter the ETC at a number of places and multiple donors can be in play; however, the underlying transport system remains the same. Electrons are ultimately transferred from donor to various redox complexes including the bc1 complex and cytochrome c, and finally to a terminal electron acceptor such as molecular oxygen in eukaryotes.[13]
The cytochrome oxidase reaction accounts for nearly 90% of all oxygen uptake in most cells.[13] Due to the large role of cytochromes within the ETC, it would be highly detrimental to the cell if any inhibitors were to be present in the organism. Cyanide and azide bind tightly to the cytochrome oxidase complex, halting electron transport and reducing the overall ATP production.[13]
Apoptosis
In all organisms, cells undergo apoptosis, or programmed cell death, by which there is an extrinsic and an intrinsic pathway. The extrinsic pathway involves an immune response by killer lymphocytes, and once the lymphocyte has been bound to the target cell, an apoptotic cascade occurs.[13] The intrinsic pathway includes cyt c, present in the intermembrane space of mitochondria. In this pathway, the presence of an apoptotic stimulus causes cyt c to be released into the cytosol. Cytochrome c in the cytosol now can be recognized and bound to various apoptotic factors, activating them and forming the apoptosome. The apoptosome recruits caspases, which are activated and result in a caspase cascade to proceed with apoptosis.[13]
Cytochrome c is required for the intrinsic apoptotic process to function properly. Such as with the electron transport chain, a mutation affecting cyt c or other structures in apoptosis could cause either an increase or a decrease in the rate of apoptosis.


