Fructose Bisphosphate Aldolase
From Proteopedia
| Line 21: | Line 21: | ||
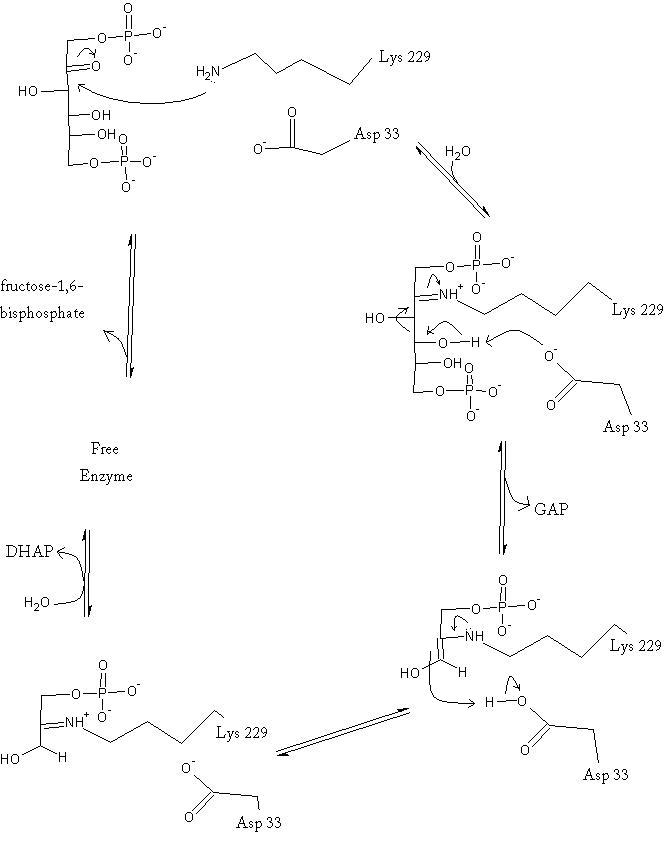
The reaction is an aldol cleavage, or otherwise termed, retro aldo condensation. Catalysis occurs first when the nucleophilic ε-amine group of Lys229 attacks the carbonyl carbon of the substrate (FBP) in its open-ring state, pushing an electron pair to the oxygen of the carbonyl. The oxygen is protonated and leaves as water as a protonated <scene name='Austin_Drake_Sandbox/Schiff_base/2'>Schiff base</scene> is produced (an imine resulting from a ketone and amine) with the open-ring form of FBP, accompanied by electrostatic stabilization from <scene name='Austin_Drake_Sandbox/Catalytic_site_w_water/5'>Asp33</scene> Aldol cleavage between C3 and C4 produces GAP and an enamine precursor to DHAP.<ref name="book" /> The cleavage is facilitated by the positive charge from the Schiff base. The subsequent electron movement, which alleviates the positive charge, also breaks the C3-C4 bond.<ref name="review" /> Tautomerization, protonation and the hydrolysis of the Schiff base produce the final product of DHAP and regenerate the enzyme. The catalysis is driven by the more favorable stability of the protonated Schiff base compared to the enolate that would appear in basic catalysis pathways.<ref name="book" /> | The reaction is an aldol cleavage, or otherwise termed, retro aldo condensation. Catalysis occurs first when the nucleophilic ε-amine group of Lys229 attacks the carbonyl carbon of the substrate (FBP) in its open-ring state, pushing an electron pair to the oxygen of the carbonyl. The oxygen is protonated and leaves as water as a protonated <scene name='Austin_Drake_Sandbox/Schiff_base/2'>Schiff base</scene> is produced (an imine resulting from a ketone and amine) with the open-ring form of FBP, accompanied by electrostatic stabilization from <scene name='Austin_Drake_Sandbox/Catalytic_site_w_water/5'>Asp33</scene> Aldol cleavage between C3 and C4 produces GAP and an enamine precursor to DHAP.<ref name="book" /> The cleavage is facilitated by the positive charge from the Schiff base. The subsequent electron movement, which alleviates the positive charge, also breaks the C3-C4 bond.<ref name="review" /> Tautomerization, protonation and the hydrolysis of the Schiff base produce the final product of DHAP and regenerate the enzyme. The catalysis is driven by the more favorable stability of the protonated Schiff base compared to the enolate that would appear in basic catalysis pathways.<ref name="book" /> | ||
| - | |||
| - | [[Image:Aldolase1.jpg]] | ||
</StructureSection> | </StructureSection> | ||
| + | [[Image:Aldolase1.jpg]] | ||
| + | |||
==Kinetics== | ==Kinetics== | ||
Revision as of 14:43, 7 November 2019
| |||||||||||
Contents |
Kinetics
Isotopic labelling has revealed the rate-determining step for the reaction. Either the carbon-carbon bond cleavage or the release of glyceraldehyde-3-phosphate comprise the slow step of the catalysis reaction; however, studies do indicate that the GAP release is likely the slowest step.[3]
It has been shown that aldolase is inhibited allosterically by oxidized glutathione, which is an oxidizing species biologically present. The glutathione oxidizes a thiol 25 angstroms from the catalytic site, which subsequently causes a drop in catalytic activity. In addition, the enzyme shows no positive cooperativity, despite being an oligomer. In fact, kinetics data actually show that the enzyme exhibits negative cooperativity. Thus the catalysis is highly compartmentalized within each subunit and binding causes little distal change of the enzymes structure.[5]
Regulation
The regulation of fructose 1,6-bisphosphate aldolase is not well understood, but the understanding is every-increasing. As it is currently observed, aldolase C appears to be regulated mainly by the gene expression--the concentration of mRNA in the cytoplasm.[6] It is also known that adenosine 3',5'-cyclicmonophosphate (cAMP) affects the expression of the gene. cAMP concentration has been positively correlated with aldolase C expression. It is believed that cAMP acts upon a section of the promotor region, distal element D, causing the transcriptional promoter, NGFI-B, to bind. Once bound, the promoter activates the transcription of the gene coding for fructose bisphosphate aldolase.[7] Given the inhibitory effects of an oxidant in the presence of aldolase, it is possible that this could be a mechanism of regulation of the enzyme. The deactivation that accompanies the oxidation of the surface thiol of Cys72 could be used intracellularly to slow the catalysis of the enzyme and regulate glycolysis.[5]
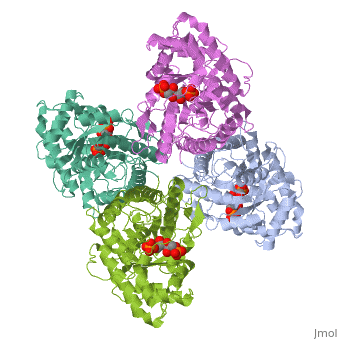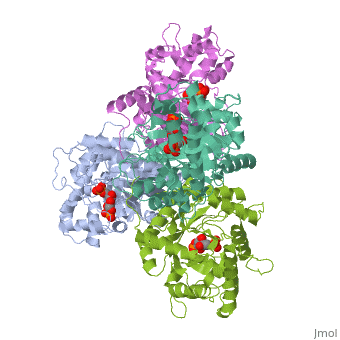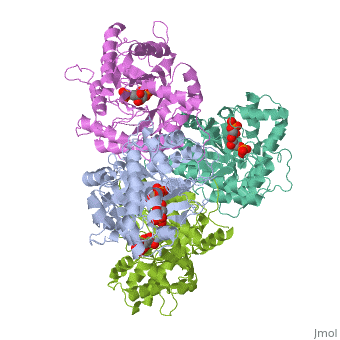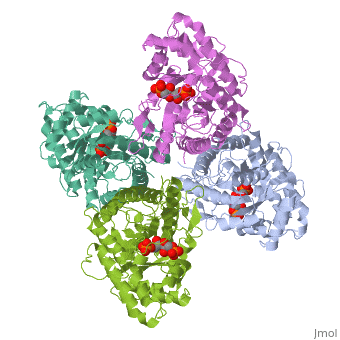
3D structures of fructose 1,6-bisphophate aldolase
Additional Resources
For additional information, see: Carbohydrate Metabolism
References
- ↑ 1.0 1.1 1.2 Voet, D, Voet, J, & Pratt, C. (2008). Fundamentals of biochemistry, third edition. Hoboken, NJ: Wiley & Sons, Inc.
- ↑ Protein: fructose-1,6-bisphosphate aldolase from human (homo sapiens), muscle isozyme. (2009). Retrieved from http://scop.mrc-lmb.cam.ac.uk
- ↑ 3.0 3.1 3.2 Gefflaut, T., B. Casimir, J. Perie, and M. Willson. "Class I Aldolases: Substrate Specificity, Mechanism, Inhibitors and Structural Aspects." Prog. Biophys. molec. Biol.. 63. (1995): 301-340.
- ↑ Dalby A, Dauter Z, Littlechild JA. Crystal structure of human muscle aldolase complexed with fructose 1,6-bisphosphate: mechanistic implications. Protein Sci. 1999 Feb;8(2):291-7. PMID:10048322
- ↑ 5.0 5.1 Sygusch, J., and Beaudry, D. "Allosteric communication in mammalian muscle aldolase." Biochem. J.. 327. (1997): 717-720.
- ↑ Paolella, G, Buono, P, Mancini, F P, Izzo, P, and Salvatore, F. "Structure and expression of mouse aldolase genes." Eur. J. Biochem.. 156. (1986): 229-235.
- ↑ Buono, P, Cassano, S, Alfieri, A, Mancini, A, and Salvatore, F. "Human aldolase C gene expression is regulated by adenosine 30,50-cyclic monophosphate (cAMP) in PC12 cells." Gene. 291. (2002): 115-121.
Proteopedia Page Contributors and Editors (what is this?)
Austin Drake, David Canner, Jacob Holt, Alexander Berchansky, Michal Harel