Phospholipase A2
From Proteopedia
(Difference between revisions)
| Line 38: | Line 38: | ||
<scene name='Journal:FLS:1/Cv/4'>Curcumin</scene> possesses anti-inflammatory activity. The binding of curcumin with PLA<sub>2</sub> was studied using X-ray crystallography. Since the electron density found in the active site did not match with curcumin, <scene name='Journal:FLS:1/Cv/5'>2-methoxycyclohexa-2-5-diene-1,4-dione (MCW)</scene> (the photo-degraded product of curcumin) <scene name='Journal:FLS:1/Cv/6'>was fitted</scene> in the unexplained electron density. To understand the <scene name='Journal:FLS:1/Cv/9'>binding mode of actual curcumin</scene>, molecular docking studies was carried out. <scene name='Journal:FLS:1/Cv/10'>Both crystallographic and docked structures were superimposed</scene> with respect to the ligand position and identified that <scene name='Journal:FLS:1/Cv/13'>curcumin is binding in the hydrophobic cavity</scene> of PLA<sub>2</sub> with a binding energy -16.81 Kcal/mol. The binding mode is in such a manner that it can prevent the entry of substrate to the hydrophobic active site. These studies indicate that curcumin can be act as an inhibitor to PLA<sub>2</sub>. | <scene name='Journal:FLS:1/Cv/4'>Curcumin</scene> possesses anti-inflammatory activity. The binding of curcumin with PLA<sub>2</sub> was studied using X-ray crystallography. Since the electron density found in the active site did not match with curcumin, <scene name='Journal:FLS:1/Cv/5'>2-methoxycyclohexa-2-5-diene-1,4-dione (MCW)</scene> (the photo-degraded product of curcumin) <scene name='Journal:FLS:1/Cv/6'>was fitted</scene> in the unexplained electron density. To understand the <scene name='Journal:FLS:1/Cv/9'>binding mode of actual curcumin</scene>, molecular docking studies was carried out. <scene name='Journal:FLS:1/Cv/10'>Both crystallographic and docked structures were superimposed</scene> with respect to the ligand position and identified that <scene name='Journal:FLS:1/Cv/13'>curcumin is binding in the hydrophobic cavity</scene> of PLA<sub>2</sub> with a binding energy -16.81 Kcal/mol. The binding mode is in such a manner that it can prevent the entry of substrate to the hydrophobic active site. These studies indicate that curcumin can be act as an inhibitor to PLA<sub>2</sub>. | ||
| - | == '''Interaction of Atropine with Phospholipase | + | == '''Interaction of Atropine with Phospholipase A2''' == |
<scene name='42/420811/Cv/1'>Atropine in complex with phospholipase A2</scene> ([[1th6]]). | <scene name='42/420811/Cv/1'>Atropine in complex with phospholipase A2</scene> ([[1th6]]). | ||
| Line 50: | Line 50: | ||
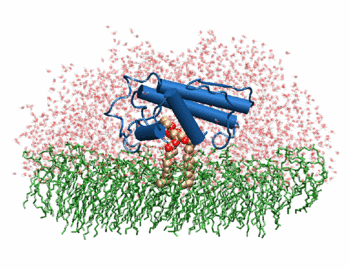
The image to the above shows the membrane-bound phospholipase A2 in blue <ref> pla2. http://www.ks.uiuc.edu/Research/smd_imd/pla2/pla2.gif </ref>. | The image to the above shows the membrane-bound phospholipase A2 in blue <ref> pla2. http://www.ks.uiuc.edu/Research/smd_imd/pla2/pla2.gif </ref>. | ||
| - | === '''Atropine in the Active Site of Phospholipase | + | === '''Atropine in the Active Site of Phospholipase A2''' === |
Atropine is an inhibitor of phospholipase 2A, and can be seen in complex with this enzyme on the left. The <scene name='Sandbox_53/Atropine_structure/1'>structure of atropine</scene> can be seen more clearly in gray using the ball-and stick representation of the drug and protein. It can also be seen in green in this <scene name='Sandbox_53/Phospholipase2a_composition/1'>space-filling model</scene>, where protein appears in brown, ligands appear in green, and solvents appear in blue. Finally, the | Atropine is an inhibitor of phospholipase 2A, and can be seen in complex with this enzyme on the left. The <scene name='Sandbox_53/Atropine_structure/1'>structure of atropine</scene> can be seen more clearly in gray using the ball-and stick representation of the drug and protein. It can also be seen in green in this <scene name='Sandbox_53/Phospholipase2a_composition/1'>space-filling model</scene>, where protein appears in brown, ligands appear in green, and solvents appear in blue. Finally, the | ||
Revision as of 12:58, 13 November 2019
| |||||||||||
3D Structures of Phospholipase A2
Updated on 13-November-2019
References
- ↑ Dennis EA. Diversity of group types, regulation, and function of phospholipase A2. J Biol Chem. 1994 May 6;269(18):13057-60. PMID:8175726
- ↑ Leitinger N, Watson AD, Hama SY, Ivandic B, Qiao JH, Huber J, Faull KF, Grass DS, Navab M, Fogelman AM, de Beer FC, Lusis AJ, Berliner JA. Role of group II secretory phospholipase A2 in atherosclerosis: 2. Potential involvement of biologically active oxidized phospholipids. Arterioscler Thromb Vasc Biol. 1999 May;19(5):1291-8. PMID:10323782
- ↑ Lapointe S, Brkovic A, Cloutier I, Tanguay JF, Arm JP, Sirois MG. Group V secreted phospholipase A2 contributes to LPS-induced leukocyte recruitment. J Cell Physiol. 2010 Jul;224(1):127-34. doi: 10.1002/jcp.22106. PMID:20232296 doi:http://dx.doi.org/10.1002/jcp.22106
- ↑ Hallstrand TS, Lai Y, Hooper KA, Oslund RC, Altemeier WA, Matute-Bello G, Gelb MH. Endogenous secreted phospholipase A2 group X regulates cysteinyl leukotrienes synthesis by human eosinophils. J Allergy Clin Immunol. 2016 Jan;137(1):268-77.e8. doi:, 10.1016/j.jaci.2015.05.026. Epub 2015 Jun 30. PMID:26139511 doi:http://dx.doi.org/10.1016/j.jaci.2015.05.026
- ↑ Platt RW, Brookhart MA, Cole SR, Westreich D, Schisterman EF. Reply to taguri and matsuyama. Stat Med. 2013 Sep 10;32(20):3592-3. doi: 10.1002/sim.5805. PMID:23943550 doi:http://dx.doi.org/10.1002/sim.5805
- ↑ Duncan RE, Sarkadi-Nagy E, Jaworski K, Ahmadian M, Sul HS. Identification and functional characterization of adipose-specific phospholipase A2 (AdPLA). J Biol Chem. 2008 Sep 12;283(37):25428-36. doi: 10.1074/jbc.M804146200. Epub 2008, Jul 9. PMID:18614531 doi:http://dx.doi.org/10.1074/jbc.M804146200
- ↑ Tjoelker LW, Wilder C, Eberhardt C, Stafforini DM, Dietsch G, Schimpf B, Hooper S, Le Trong H, Cousens LS, Zimmerman GA, Yamada Y, McIntyre TM, Prescott SM, Gray PW. Anti-inflammatory properties of a platelet-activating factor acetylhydrolase. Nature. 1995 Apr 6;374(6522):549-53. PMID:7700381 doi:http://dx.doi.org/10.1038/374549a0
- ↑ Quach ND, Arnold RD, Cummings BS. Secretory phospholipase A2 enzymes as pharmacological targets for treatment of disease. Biochem Pharmacol. 2014 Aug 15;90(4):338-48. doi: 10.1016/j.bcp.2014.05.022. Epub, 2014 Jun 4. PMID:24907600 doi:http://dx.doi.org/10.1016/j.bcp.2014.05.022
- ↑ Tellis CC, Tselepis AD. The role of lipoprotein-associated phospholipase A2 in atherosclerosis may depend on its lipoprotein carrier in plasma. Biochim Biophys Acta. 2009 May;1791(5):327-38. doi: 10.1016/j.bbalip.2009.02.015. PMID:19272461 doi:http://dx.doi.org/10.1016/j.bbalip.2009.02.015
- ↑ Singh N, Jabeen T, Sharma S, Somvanshi RK, Dey S, Srinivasan A, Singh TP. Specific binding of non-steroidal anti-inflammatory drugs (NSAIDs) to phospholipase A2: structure of the complex formed between phospholipase A2 and diclofenac at 2.7 A resolution. Acta Crystallogr D Biol Crystallogr. 2006 Apr;62(Pt 4):410-6. Epub 2006, Mar 18. PMID:16552142 doi:10.1107/S0907444906003660
- ↑ Crystal structure of porcine pancreatic phospholipase a2 in complex with 2-methoxycyclohexa-2-5-diene-1,4-dione. Dileep KV, Tintu I, Mandal PK, Karthe P, Haridas M, Sadasivan C. Frontiers In Life Sci. (2012) doi:http://dx.doi.org/10.1080/21553769.2012.689262
- ↑ Kumar, Jainendra; Bala, Priti; Vihwal, Preeti. Analysis of Interaction of atropine with phospholipase A2 (1th6.pdb). Department of Botany and Biotechnlogy, College of Commerce, Patna, India.
- ↑ Phospholipase A2. http://www.worldlingo.com/ma/enwiki/en/Phospholipase_A2
- ↑ Phospholipase A2. http://www.worldlingo.com/ma/enwiki/en/Phospholipase_A2
- ↑ Phospholipase A2. http://www.worldlingo.com/ma/enwiki/en/Phospholipase_A2
- ↑ pla2.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky