We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Phosphoglycerate Mutase
From Proteopedia
(Difference between revisions)
| Line 31: | Line 31: | ||
== Phosphoglycerate Mutase Deficiency == | == Phosphoglycerate Mutase Deficiency == | ||
When phosphoglycerate mutase has a genetic defect, it results in a muscle disease that interferes with the processing of carbohydrates. The onset can occur anywhere from childhood to adulthood. The inheritance pattern is autosomal recessive. <ref>http://www.mda.org/disease/pgam.html</ref> Phosphoglycerate mutase deficicncy patients may experience CNS symptoms such as mental retardation and seizures. Certain individuals may experience a purely myopathic syndrome with progressive proximal muscle weakness and incidents of myoglobinuria, exercise intolerance, may become easy fatigued with cramps and urine discoloration. Diagnosing this deficiency can be done with Laboratory tests that demonstrate and increased serum CK level. Or there are diagnostic tests available that test for the absence of the enzyme. Also, muscle pathology of this deficiency shows subsarcolemmal glycogen ± tubular combinations. <ref>http://disability.ucdavis.edu/disease_deatails.php?id=45</ref> | When phosphoglycerate mutase has a genetic defect, it results in a muscle disease that interferes with the processing of carbohydrates. The onset can occur anywhere from childhood to adulthood. The inheritance pattern is autosomal recessive. <ref>http://www.mda.org/disease/pgam.html</ref> Phosphoglycerate mutase deficicncy patients may experience CNS symptoms such as mental retardation and seizures. Certain individuals may experience a purely myopathic syndrome with progressive proximal muscle weakness and incidents of myoglobinuria, exercise intolerance, may become easy fatigued with cramps and urine discoloration. Diagnosing this deficiency can be done with Laboratory tests that demonstrate and increased serum CK level. Or there are diagnostic tests available that test for the absence of the enzyme. Also, muscle pathology of this deficiency shows subsarcolemmal glycogen ± tubular combinations. <ref>http://disability.ucdavis.edu/disease_deatails.php?id=45</ref> | ||
| + | |||
| + | ==3D structures of phosphoglycerate mutase== | ||
| + | [[Phosphoglycerate mutase 3D structures]] | ||
| + | |||
</StructureSection> | </StructureSection> | ||
==3D structures of phosphoglycerate mutase== | ==3D structures of phosphoglycerate mutase== | ||
| Line 40: | Line 44: | ||
**[[3o0t]], [[3mxo]] – hPGM5 residues 90-289 – human<br /> | **[[3o0t]], [[3mxo]] – hPGM5 residues 90-289 – human<br /> | ||
| - | **[[1yfk]], [[1yjx]], [[4gpi]], [[4gpz]] – hPGM1<br /> | + | **[[1yfk]], [[1yjx]], [[4gpi]], [[4gpz]], [[5zs7]] – hPGM1<br /> |
| + | **[[6h26]] - PGM – rabbit<br /> | ||
| + | **[[1fzt]] – PGM – Fission yeast – NMR<br /> | ||
| + | **[[5pgm]], [[4pgm]], [[3pgm]] – yPGM – yeast<br /> | ||
**[[3ezn]] – BpPGM – ''Burkholderia pseudomallei''<br /> | **[[3ezn]] – BpPGM – ''Burkholderia pseudomallei''<br /> | ||
**[[3d8h]] – PGM – ''Cryptosporidium parvum''<br /> | **[[3d8h]] – PGM – ''Cryptosporidium parvum''<br /> | ||
| Line 48: | Line 55: | ||
**[[1rii]] – PGM – ''Mycobacteriumtuberculosis''<br /> | **[[1rii]] – PGM – ''Mycobacteriumtuberculosis''<br /> | ||
**[[1e58]] – EcPGM – ''Escherichia coli''<br /> | **[[1e58]] – EcPGM – ''Escherichia coli''<br /> | ||
| - | **[[1fzt]] – PGM – Fission yeast – NMR<br /> | ||
| - | **[[5pgm]], [[4pgm]], [[3pgm]] – yPGM – yeast<br /> | ||
**[[5vve]] - PGM – ''Naegleria fowleri''<br /> | **[[5vve]] - PGM – ''Naegleria fowleri''<br /> | ||
*Phosphoglycerate mutase binary complexes | *Phosphoglycerate mutase binary complexes | ||
| + | **[[5y2i]], [[5y35]], [[5y64]], [[5y65]], [[5zrm]], [[5zs8]] – hPGM1 + inhibitor<br /> | ||
| + | **[[5y2u]] – hPGM1 + alizarin red<br /> | ||
| + | **[[1qhf]] – yPGM + PGA <br /> | ||
| + | **[[1bq3]] – yPGM + inositol hexakisphosphate<br /> | ||
| + | **[[1bq4]] – yPGM + benzene hexacarboxylate<br /> | ||
**[[3lnt]] – BpPGM + malonic acid <br /> | **[[3lnt]] – BpPGM + malonic acid <br /> | ||
**[[3gw8]] – BpPGM + glycerol + VO4<BR /> | **[[3gw8]] – BpPGM + glycerol + VO4<BR /> | ||
| Line 61: | Line 71: | ||
**[[1eqj]], [[1ejj]] – BsPGM + PGA – ''Bacillus stearothermophilus''<br /> | **[[1eqj]], [[1ejj]] – BsPGM + PGA – ''Bacillus stearothermophilus''<br /> | ||
**[[1e59]] – EcPGM + VO4<BR /> | **[[1e59]] – EcPGM + VO4<BR /> | ||
| - | **[[1qhf]] – yPGM + PGA <br /> | ||
| - | **[[1bq3]] – yPGM + inositol hexakisphosphate<br /> | ||
| - | **[[1bq4]] – yPGM + benzene hexacarboxylate<br /> | ||
| - | **[[5y2i]], [[5y35]] – hPGM1 + inhibitor<br /> | ||
| - | **[[5y2u]] – hPGM1 + alizarin red<br /> | ||
*2,3-bisphosphoglycerate-independent phosphoglycerate mutase | *2,3-bisphosphoglycerate-independent phosphoglycerate mutase | ||
| Line 76: | Line 81: | ||
**[[4my4]] – SaPGM – ''Staphylococcus aureus''<br /> | **[[4my4]] – SaPGM – ''Staphylococcus aureus''<br /> | ||
**[[3idd]], [[3kd8]] – TaBIPGM – ''Thermoplasma acidophilum''<br /> | **[[3idd]], [[3kd8]] – TaBIPGM – ''Thermoplasma acidophilum''<br /> | ||
| - | **[[3nvl]] – | + | **[[3nvl]] – PGM - ''Trypanosoma brucei''<br /> |
| - | **[[2ify]] – | + | **[[2ify]] – PGM – ''Bacillus anthracis''<br /> |
| - | **[[2zkt]] – | + | **[[2zkt]] – PGM – ''Pyrococcus horikoshii''<br /> |
*2,3-bisphosphoglycerate-independent phosphoglycerate mutase complexes | *2,3-bisphosphoglycerate-independent phosphoglycerate mutase complexes | ||
| - | **[[4nwj]] – SaPGM + 3PG<br /> | ||
| - | **[[4nwx]], [[4qax]] – SaPGM + 2PG<br /> | ||
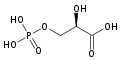
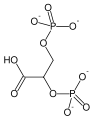
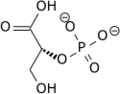
**[[2a9j]], [[2h4x]], [[2h52]] – hPGM + 3PG<br /> | **[[2a9j]], [[2h4x]], [[2h52]] – hPGM + 3PG<br /> | ||
**[[2f90]] - hPGM + 3PG + AlF4<br /> | **[[2f90]] - hPGM + 3PG + AlF4<br /> | ||
**[[2h4z]], [[2hhj]] – hPGM + 2,3-BGP<br /> | **[[2h4z]], [[2hhj]] – hPGM + 2,3-BGP<br /> | ||
| + | **[[4nwj]] – SaPGM + 3PG<br /> | ||
| + | **[[4nwx]], [[4qax]] – SaPGM + 2PG<br /> | ||
**[[1ejj]] - GsPGM + 3PG – ''Geobacillus stearothermophilus''<br /> | **[[1ejj]] - GsPGM + 3PG – ''Geobacillus stearothermophilus''<br /> | ||
**[[1eqj]] - GsPGM + 2PG<br /> | **[[1eqj]] - GsPGM + 2PG<br /> | ||
Revision as of 08:42, 18 November 2019
| |||||||||||
3D structures of phosphoglycerate mutase
Updated on 18-November-2019
Additional Resources
For additional information, please see: Carbohydrate Metabolism
References
- ↑ Crowhurst GS, Dalby AR, Isupov MN, Campbell JW, Littlechild JA. Structure of a phosphoglycerate mutase:3-phosphoglyceric acid complex at 1.7 A. Acta Crystallogr D Biol Crystallogr. 1999 Nov;55(Pt 11):1822-6. PMID:10531478
- ↑ http://disability.ucdavis.edu/disease_deatails.php?id=45
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 S., Winn I., Fothergill A. L., Harkins N. R., and Watson C. H. "Structure and Activity of Phosphoglycerate Mutase." Sciences 293.1063 (1981): 121-30. Print.
- ↑ "Phosphoglycerate mutase -." Wikipedia, the free encyclopedia. Web. 27 Feb. 2010. <http://en.wikipedia.org/wiki/Phosphoglycerate_mutase>.
- ↑ 5.0 5.1 5.2 Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry Life at the Molecular Level. New York: John Wiley & Sons, 2008. Print.
- ↑ Rose, Z.B. (1980) Adv. Enzymol. Relat. Areas Mol. Biol. 51, 211-253
- ↑ Rigden, D. J.; Walter, R. A.; Phillips, S. E. V.; Fothergill-Gilmore, L. A.Polyanionic inhibitors of phosphoglycerate mutase: combined structural and biochemical analysis J. Mol. Biol. 1999, 289, 691– 699
- ↑ McAleese, S.M., Fothergill-Gilmore, L.A.&Dixon, H.B.F. (1985) Biochem. J. 230, 535-542
- ↑ http://www.mda.org/disease/pgam.html
- ↑ http://disability.ucdavis.edu/disease_deatails.php?id=45
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Robert Trahin, Xuan Loi, David Canner, Christopher Vachon, Allie Paton