We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Porin
From Proteopedia
(Difference between revisions)
| Line 31: | Line 31: | ||
The approach and algorithm used here requires less CPU power and time than MD, and makes it possible to conduct molecular studies of large molecules with known atomic structure at shorter time. | The approach and algorithm used here requires less CPU power and time than MD, and makes it possible to conduct molecular studies of large molecules with known atomic structure at shorter time. | ||
| + | |||
| + | == 3D structures of Porin == | ||
| + | [[Porin 3D structures]] | ||
| + | |||
</StructureSection> | </StructureSection> | ||
| Line 65: | Line 69: | ||
**[[2j4u]] – EcOmpC+lactotransferrin fragment<br /> | **[[2j4u]] – EcOmpC+lactotransferrin fragment<br /> | ||
**[[3upg]], [[3uu2]] – SeOmpC <br /> | **[[3upg]], [[3uu2]] – SeOmpC <br /> | ||
| - | **[[5fvn]] – | + | **[[5fvn]] – EncOmpC – ''Enterobacter cloacae''<br /> |
*OmpF | *OmpF | ||
| Line 80: | Line 84: | ||
**[[3nsg]] – StOmpF - ''Salmonella typhimurium''<br /> | **[[3nsg]] – StOmpF - ''Salmonella typhimurium''<br /> | ||
**[[4kr4]], [[4kr8]], [[4kra]] – StOmpF + antibiotics<br /> | **[[4kr4]], [[4kr8]], [[4kra]] – StOmpF + antibiotics<br /> | ||
| + | **[[6ene]] – EncOmpF<br /> | ||
*OmpG | *OmpG | ||
| Line 89: | Line 94: | ||
*OmpK | *OmpK | ||
| - | **[[1osm]] – KpOmpK36 <br /> | + | **[[1osm]], [[6rck]] – KpOmpK36 <br /> |
| - | **[[5nup]] – KpOmpK36 + ABC | + | **[[6rcp]] – KpOmpK36 (mutant)<br /> |
| + | **[[5nup]] – KpOmpK36 + ABC transporter permease<br /> | ||
*OmpM | *OmpM | ||
| Line 109: | Line 115: | ||
**[[2fgr]] - DaOmp32 – ''Delftia acidovorans''<br /> | **[[2fgr]] - DaOmp32 – ''Delftia acidovorans''<br /> | ||
| - | **[[1e54]] – Omp32+sulfate – ''Comamonas acidovorans''<br /> | ||
**[[2fgq]] – DaOmp32+malate<br /> | **[[2fgq]] – DaOmp32+malate<br /> | ||
| + | **[[1e54]] – Omp32+sulfate – ''Comamonas acidovorans''<br /> | ||
| + | |||
| + | *Omp35 | ||
| + | |||
| + | **[[5o78]] – Omp35 – ''Enterobacter aerogenes''<br /> | ||
*OprB | *OprB | ||
| Line 197: | Line 207: | ||
**[[5nxu]] – PsOmp 1 + maltose<br /> | **[[5nxu]] – PsOmp 1 + maltose<br /> | ||
**[[4d65]] – PsOmp 2<br /> | **[[4d65]] – PsOmp 2<br /> | ||
| - | **[[4fuv]], [[ | + | **[[4fuv]], [[5dl7]] – Omp – ''Acinetobacter baumannii''<br /> |
*Sugar-specific porin | *Sugar-specific porin | ||
Revision as of 10:35, 25 November 2019
| |||||||||||
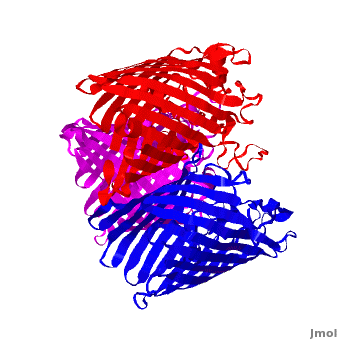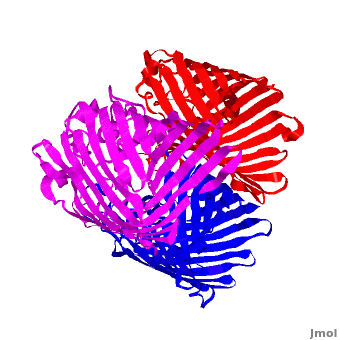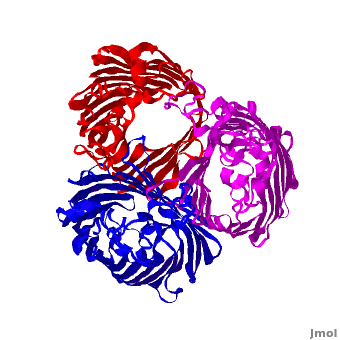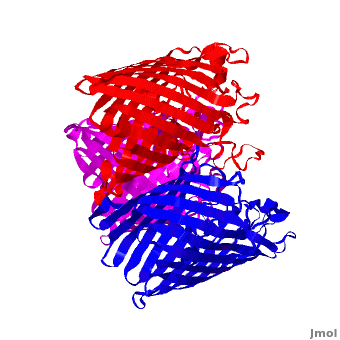
3D structures of Porin
Updated on 25-November-2019
Voltage-Dependent Anion Channel
See Ion channels
References
- ↑ Shoshan-Barmatz V, Israelson A, Brdiczka D, Sheu SS. The voltage-dependent anion channel (VDAC): function in intracellular signalling, cell life and cell death. Curr Pharm Des. 2006;12(18):2249-70. PMID:16787253
- ↑ Van Gelder P, Dumas F, Bartoldus I, Saint N, Prilipov A, Winterhalter M, Wang Y, Philippsen A, Rosenbusch JP, Schirmer T. Sugar transport through maltoporin of Escherichia coli: role of the greasy slide. J Bacteriol. 2002 Jun;184(11):2994-9. PMID:12003940
- ↑ Basle A, Rummel G, Storici P, Rosenbusch JP, Schirmer T. Crystal structure of osmoporin OmpC from E. coli at 2.0 A. J Mol Biol. 2006 Oct 6;362(5):933-42. Epub 2006 Aug 3. PMID:16949612 doi:10.1016/j.jmb.2006.08.002
- ↑ Besya AB, Mobasheri H, Ejtehadi MR. Gating and conduction of nano-channel forming proteins: a computational approach. J Biomol Struct Dyn. 2012 Aug 28. PMID:22928968 doi:10.1080/07391102.2012.712460
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Karsten Theis, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky