1a0a
From Proteopedia
| Line 1: | Line 1: | ||
[[Image:1a0a.gif|left|200px]] | [[Image:1a0a.gif|left|200px]] | ||
| - | + | <!-- | |
| - | + | The line below this paragraph, containing "STRUCTURE_1a0a", creates the "Structure Box" on the page. | |
| - | + | You may change the PDB parameter (which sets the PDB file loaded into the applet) | |
| - | + | or the SCENE parameter (which sets the initial scene displayed when the page is loaded), | |
| - | | | + | or leave the SCENE parameter empty for the default display. |
| - | | | + | --> |
| - | + | {{STRUCTURE_1a0a| PDB=1a0a | SCENE= }} | |
| - | + | ||
| - | + | ||
| - | }} | + | |
'''PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4/DNA COMPLEX''' | '''PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4/DNA COMPLEX''' | ||
| Line 33: | Line 30: | ||
[[Category: Shimizu, T.]] | [[Category: Shimizu, T.]] | ||
[[Category: Toumoto, A.]] | [[Category: Toumoto, A.]] | ||
| - | [[Category: | + | [[Category: Basic helix loop helix]] |
| - | [[Category: | + | [[Category: Transcription factor]] |
| - | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Fri May 2 09:36:57 2008'' | |
| - | + | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | |
Revision as of 06:36, 2 May 2008
| |||||||||
| 1a0a, resolution 2.80Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4/DNA COMPLEX
Overview
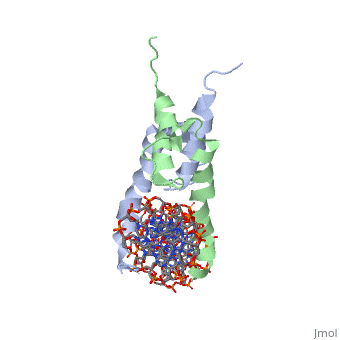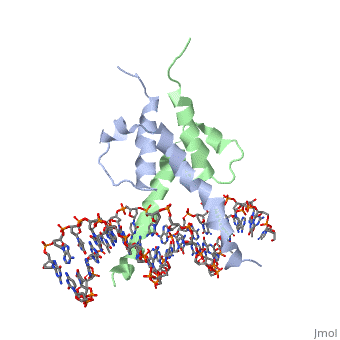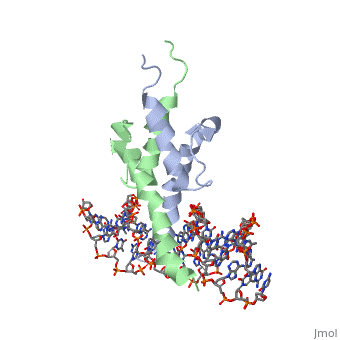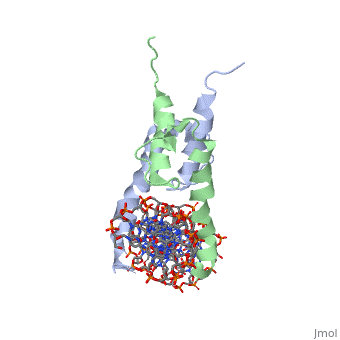
The crystal structure of a DNA-binding domain of PHO4 complexed with DNA at 2.8 A resolution revealed that the domain folds into a basic-helix-loop-helix (bHLH) motif with a long but compact loop that contains a short alpha-helical segment. This helical structure positions a tryptophan residue into an aromatic cluster so as to make the loop compact. PHO4 binds to DNA as a homodimer with direct reading of both the core E-box sequence CACGTG and its 3'-flanking bases. The 3'-flanking bases GG are recognized by Arg2 and His5. The residues involved in the E-box recognition are His5, Glu9 and Arg13, as already reported for bHLH/Zip proteins MAX and USF, and are different from those recognized by bHLH proteins MyoD and E47, although PHO4 is a bHLH protein.
About this Structure
1A0A is a Single protein structure of sequence from Saccharomyces cerevisiae. Full crystallographic information is available from OCA.
Reference
Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition., Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T, EMBO J. 1997 Aug 1;16(15):4689-97. PMID:9303313 Page seeded by OCA on Fri May 2 09:36:57 2008