We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1568
From Proteopedia
(Difference between revisions)
| Line 24: | Line 24: | ||
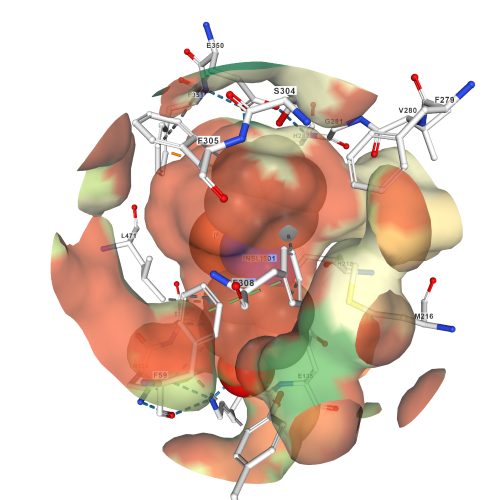
The <scene name='82/823092/Catalytic_triad/2'>catalytic triad</scene> of the binding site consists of Phe59, Tyr101, and Lys134 that contact the 4-hydroxyphenyl portion of the substrate. | The <scene name='82/823092/Catalytic_triad/2'>catalytic triad</scene> of the binding site consists of Phe59, Tyr101, and Lys134 that contact the 4-hydroxyphenyl portion of the substrate. | ||
| - | Important <scene name='82/823092/Active_site_interactions/1'>interactions in the active site</scene>. | + | Important <scene name='82/823092/Active_site_interactions/1'>interactions in the active site</scene> are shown. Green indicates hydrophobic interactions, blue indicates hydrogen bonding interactions, and the orange nucleotides are specific histidines that support and interact with the metal ion (Fe) in the pocket. These two histidines also contribute to hydrogen bonds in the area. |
== '''Energy Transformation''' == | == '''Energy Transformation''' == | ||
Revision as of 21:03, 30 November 2019
| This Sandbox is Reserved from Aug 26 through Dec 12, 2019 for use in the course CHEM 351 Biochemistry taught by Bonnie_Hall at the Grand View University, Des Moines, USA. This reservation includes Sandbox Reserved 1556 through Sandbox Reserved 1575. |
To get started:
More help: Help:Editing |
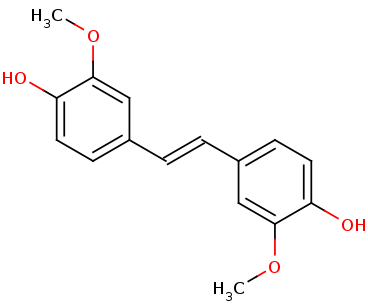
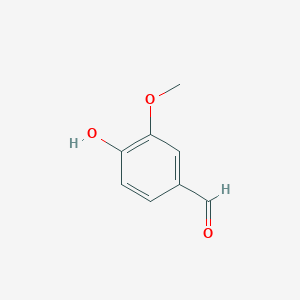
Lignostilbene-α,ß-dioxygenase A structural features and important functional residues
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Kuatsjah E, Verstraete MM, Kobylarz MJ, Liu AKN, Murphy MEP, Eltis LD. Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase. J Biol Chem. 2019 Jul 10. pii: RA119.009428. doi: 10.1074/jbc.RA119.009428. PMID:31292192 doi:http://dx.doi.org/10.1074/jbc.RA119.009428