Ribonuclease A Catalysis
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
<StructureSection load='7rsa' size='350' side='right' caption='' scene='Sandbox_Reserved_193/Rnasei_a/1 ' pspeed='8'> | <StructureSection load='7rsa' size='350' side='right' caption='' scene='Sandbox_Reserved_193/Rnasei_a/1 ' pspeed='8'> | ||
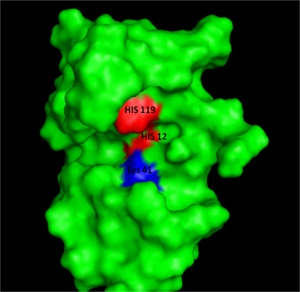
[[Image:RNaseAIII.png|300px|left|thumb|Figure I: Bovine Ribonuclease A. Colored residues are representative of amino acids important to both the acid base catalysis (Red: His12 and 119) and stabilization of the transition state (Blue: Lys41). Figure generated via ''Pymol'' ]] | [[Image:RNaseAIII.png|300px|left|thumb|Figure I: Bovine Ribonuclease A. Colored residues are representative of amino acids important to both the acid base catalysis (Red: His12 and 119) and stabilization of the transition state (Blue: Lys41). Figure generated via ''Pymol'' ]] | ||
| - | + | {{Clear}} | |
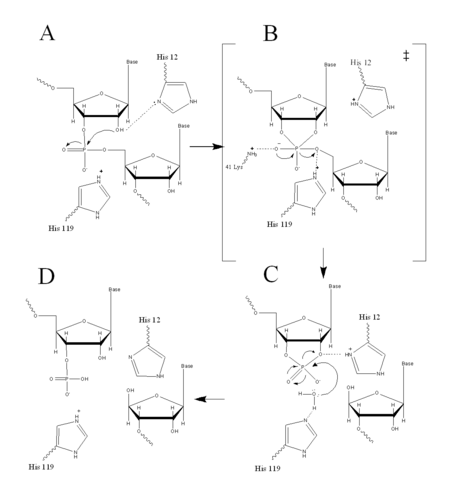
===Acid Base Catalysis=== | ===Acid Base Catalysis=== | ||
In organic chemistry acid/base catalysis is the addition of an acid or base to accelerate a chemical reaction. Ribonuclease A, (RNase A), also uses acid/base catatalysis to chemically change its substrates. Acidic or basic residues of the enzyme transfer protons to or from the reactant in order to stabilize the developing charges in the transition state. The transfer of protons usually creates better leaving groups, making the reaction more energetically favorable. Histidine is a very common amino acid residue involved in cataylsis, as histidine has a pKa value close to neutral, (pKa=6); therefore, histidine can both accept and donate protons at physiological pH. | In organic chemistry acid/base catalysis is the addition of an acid or base to accelerate a chemical reaction. Ribonuclease A, (RNase A), also uses acid/base catatalysis to chemically change its substrates. Acidic or basic residues of the enzyme transfer protons to or from the reactant in order to stabilize the developing charges in the transition state. The transfer of protons usually creates better leaving groups, making the reaction more energetically favorable. Histidine is a very common amino acid residue involved in cataylsis, as histidine has a pKa value close to neutral, (pKa=6); therefore, histidine can both accept and donate protons at physiological pH. | ||
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Wlodrawer, A., Svensson, L., Sjohin, L., Gilliland, G. Structure of Phosphate-Free Ribonuclease A Refined at 1.26A. Biochemistry:(1988) Vol. 27 pp. 2705-2717
- ↑ 2.0 2.1 2.2 2.3 Raines, R. Ribonuclease A. Chemistry Review: (1998) Vol. 98 pp. 1045-1068
- ↑ 3.0 3.1 Vicentini,A., Protein Chemical and Kinetic Characterization of Recombinant Porcine Ribonuclease Inhibitor Expressed in Saccharomyces cerevisae. Biochemistry: (1996) Vol. 29, pp.8827-8834
- ↑ 4.0 4.1 Turcotte, R., Raines, R., Interactions of Onconase with Human Ribonuclease Inhibitor. Biochemical Biophysical Research Communities:(2008) Vol. 377, Iss. 4, pp. 512-414
Related Sites
http://en.wikipedia.org/wiki/RNase_A
http://en.wikipedia.org/wiki/Ribonuclease_inhibitor
Proteopedia Page Contributors and Editors (what is this?)
Nathan Clarke, David Canner, Alexander Berchansky, R. Jeremy Johnson, OCA