We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1568
From Proteopedia
(Difference between revisions)
| Line 17: | Line 17: | ||
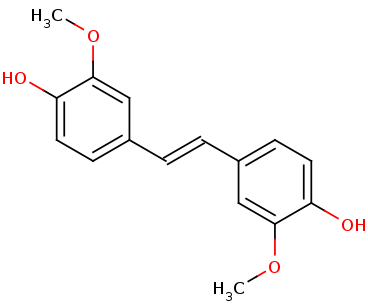
When you look at the <scene name='82/823092/Spacefill_lsda/1'>spacefill view</scene> of the protein dimer you see that the binding pocket accessibility is very restrictive. | When you look at the <scene name='82/823092/Spacefill_lsda/1'>spacefill view</scene> of the protein dimer you see that the binding pocket accessibility is very restrictive. | ||
| - | [[Image:spacefill hydrophobicity.png]][[Image:ligand hydrophobicity.png]] | ||
| - | <scene name='82/823092/Hydrophobic_spacefill/1'>Hydrophobicity-focused</scene> view of the protein. | + | This is a <scene name='82/823092/Hydrophobic_spacefill/1'>Hydrophobicity-focused</scene> view of the protein. Overall, there doesn't seem to be any dominant hydrophobic or hydrophillic regions of the protein. |
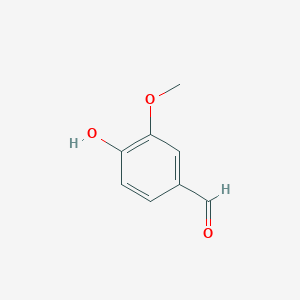
The <scene name='82/823092/Catalytic_triad/2'>catalytic triad</scene> of the binding site consists of Phe59, Tyr101, and Lys134 that interact with the 4-hydroxyphenyl portion of the substrate. The triad importance was tested with specific mutations. A F59H mutation led to 3% efficiency comparable to wildtype LsdA. A Y101F mutation led to 20% efficiency comparable to wildtype LsdA. And a K134M mutation showed no discernible lignostilbene cleavage activity <ref>PMID 31292192</ref>. | The <scene name='82/823092/Catalytic_triad/2'>catalytic triad</scene> of the binding site consists of Phe59, Tyr101, and Lys134 that interact with the 4-hydroxyphenyl portion of the substrate. The triad importance was tested with specific mutations. A F59H mutation led to 3% efficiency comparable to wildtype LsdA. A Y101F mutation led to 20% efficiency comparable to wildtype LsdA. And a K134M mutation showed no discernible lignostilbene cleavage activity <ref>PMID 31292192</ref>. | ||
Revision as of 23:45, 8 December 2019
| This Sandbox is Reserved from Aug 26 through Dec 12, 2019 for use in the course CHEM 351 Biochemistry taught by Bonnie_Hall at the Grand View University, Des Moines, USA. This reservation includes Sandbox Reserved 1556 through Sandbox Reserved 1575. |
To get started:
More help: Help:Editing |
Lignostilbene-α,ß-dioxygenase A structural features and important functional residues
| |||||||||||
References
- ↑ Kuatsjah E, Verstraete MM, Kobylarz MJ, Liu AKN, Murphy MEP, Eltis LD. Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase. J Biol Chem. 2019 Jul 10. pii: RA119.009428. doi: 10.1074/jbc.RA119.009428. PMID:31292192 doi:http://dx.doi.org/10.1074/jbc.RA119.009428
- ↑ Kuatsjah E, Verstraete MM, Kobylarz MJ, Liu AKN, Murphy MEP, Eltis LD. Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase. J Biol Chem. 2019 Jul 10. pii: RA119.009428. doi: 10.1074/jbc.RA119.009428. PMID:31292192 doi:http://dx.doi.org/10.1074/jbc.RA119.009428
- ↑ Kuatsjah E, Verstraete MM, Kobylarz MJ, Liu AKN, Murphy MEP, Eltis LD. Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase. J Biol Chem. 2019 Jul 10. pii: RA119.009428. doi: 10.1074/jbc.RA119.009428. PMID:31292192 doi:http://dx.doi.org/10.1074/jbc.RA119.009428