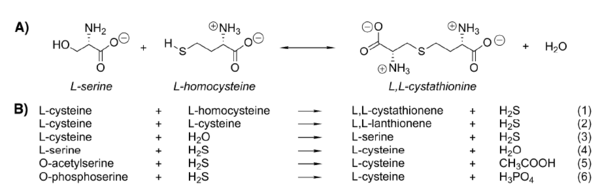
Cystathionine β-synthase
From Proteopedia
| Line 79: | Line 79: | ||
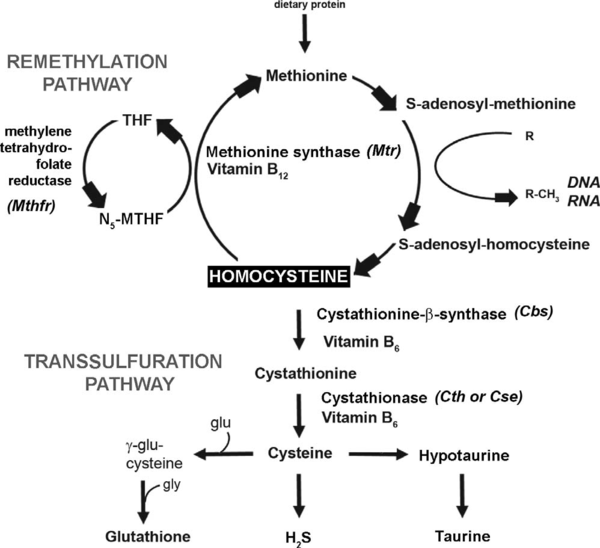
A second popular hypothesis suggests that eHcy affects the control of biologically important methylation reactions by causing a build-up of S-adenosyl-L-homocysteine (AdoHcy) which is a competitive inhibitor of S-adenosyl-L-methionine (AdoMet) binding for methyltransferase enzymes. As methyltransferases are involved in a variety of important biological processes, inhibition of this class of enzymes could have extremely diverse effects on the organism.<ref>PMID:15890029</ref> | A second popular hypothesis suggests that eHcy affects the control of biologically important methylation reactions by causing a build-up of S-adenosyl-L-homocysteine (AdoHcy) which is a competitive inhibitor of S-adenosyl-L-methionine (AdoMet) binding for methyltransferase enzymes. As methyltransferases are involved in a variety of important biological processes, inhibition of this class of enzymes could have extremely diverse effects on the organism.<ref>PMID:15890029</ref> | ||
</StructureSection> | </StructureSection> | ||
| + | |||
| + | ==3D structures of Cystathionine β-synthase== | ||
| + | |||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | |||
| + | [[4l3v]], [[4coo]] – hCBS + PLP – human<br /> | ||
| + | [[5mms]], [[4l0d]], [[4l28]], [[4l27]] – hCBS (mutant) + PLP <br /> | ||
| + | [[1jbq]] – hCBS 1-412 (mutant) + PLP <br /> | ||
| + | [[1m54]] – hCBS 44-406 (mutant) + PLP <br /> | ||
| + | [[4pcu]] – hCBS + PLP + SAM<br /> | ||
| + | [[4uuu]] – hCBS regulatory domain 406-547 + SAM<br /> | ||
| + | [[3pc2]] – DmCBS + PLP – ''Drosophila melanogaster''<br /> | ||
| + | [[3pc3]] – DmCBS + PLP + aminoacrylate<br /> | ||
| + | [[3pc4]] – DmCBS + PLP + Ser<br /> | ||
| + | [[5ohx]] – CBS + PLP – bee<br /> | ||
| + | [[6c2h]] – yCBS catalytic domain 1-353 + PLP – yeast<br /> | ||
| + | [[6c2q]] – yCBS catalytic domain + PLP-Ser<br /> | ||
| + | [[6c2z]] – yCBS catalytic domain + PLP-aminoacrylate<br /> | ||
| + | [[6c4p]] – yCBS catalytic domain + PMP<br /> | ||
| + | [[5b1h]] – LpCBS + PLP derivative – ''Lactobacillus plantarum''<br /> | ||
| + | [[5b1i]] – LpCBS (mutant) + Met derivative<br /> | ||
| + | [[5xw3]] – CBS – ''Bacillus anthracis''<br /> | ||
| + | [[6ahi]] – CBS + PLP derivative + Met – ''Helicobacter pylori''<br /> | ||
| + | |||
== Credits == | == Credits == | ||
Article created as an [https://is.cuni.cz/studium/predmety/index.php?id=b0728408dfab8c4e0250521ab9988dd8&tid=&do=predmet&kod=MB151P117&skr=2018 Structural biology of the cell] assignment at the Faculty of Science, Charles University, Prague, Czech Republic. | Article created as an [https://is.cuni.cz/studium/predmety/index.php?id=b0728408dfab8c4e0250521ab9988dd8&tid=&do=predmet&kod=MB151P117&skr=2018 Structural biology of the cell] assignment at the Faculty of Science, Charles University, Prague, Czech Republic. | ||
| Line 85: | Line 109: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | |||
| + | [[Category:Topic Page]] | ||
Revision as of 07:53, 9 January 2020
Contents |
3D Structure of Human Cystathionine β-synthase (4coo)
| |||||||||||
3D structures of Cystathionine β-synthase
Updated on 09-January-2020
4l3v, 4coo – hCBS + PLP – human
5mms, 4l0d, 4l28, 4l27 – hCBS (mutant) + PLP
1jbq – hCBS 1-412 (mutant) + PLP
1m54 – hCBS 44-406 (mutant) + PLP
4pcu – hCBS + PLP + SAM
4uuu – hCBS regulatory domain 406-547 + SAM
3pc2 – DmCBS + PLP – Drosophila melanogaster
3pc3 – DmCBS + PLP + aminoacrylate
3pc4 – DmCBS + PLP + Ser
5ohx – CBS + PLP – bee
6c2h – yCBS catalytic domain 1-353 + PLP – yeast
6c2q – yCBS catalytic domain + PLP-Ser
6c2z – yCBS catalytic domain + PLP-aminoacrylate
6c4p – yCBS catalytic domain + PMP
5b1h – LpCBS + PLP derivative – Lactobacillus plantarum
5b1i – LpCBS (mutant) + Met derivative
5xw3 – CBS – Bacillus anthracis
6ahi – CBS + PLP derivative + Met – Helicobacter pylori
Credits
Article created as an Structural biology of the cell assignment at the Faculty of Science, Charles University, Prague, Czech Republic.
Assignment authors: Jana Křivková, Zdeňka Mauerová, Jan Hamalčík
References
- ↑ Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein. EMBO J. 2001 Aug 1;20(15):3910-6. PMID:11483494 doi:http://dx.doi.org/10.1093/emboj/20.15.3910
- ↑ Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein. EMBO J. 2001 Aug 1;20(15):3910-6. PMID:11483494 doi:http://dx.doi.org/10.1093/emboj/20.15.3910
- ↑ Meier M, Oliveriusova J, Kraus JP, Burkhard P. Structural insights into mutations of cystathionine beta-synthase. Biochim Biophys Acta. 2003 Apr 11;1647(1-2):206-13. PMID:12686134
- ↑ Tu Y, Kreinbring CA, Hill M, Liu C, Petsko GA, McCune CD, Berkowitz DB, Liu D, Ringe D. Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time. Biochemistry. 2018 Apr 13. doi: 10.1021/acs.biochem.8b00092. PMID:29630349 doi:http://dx.doi.org/10.1021/acs.biochem.8b00092
- ↑ Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein. EMBO J. 2001 Aug 1;20(15):3910-6. PMID:11483494 doi:http://dx.doi.org/10.1093/emboj/20.15.3910
- ↑ Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein. EMBO J. 2001 Aug 1;20(15):3910-6. PMID:11483494 doi:http://dx.doi.org/10.1093/emboj/20.15.3910
- ↑ Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein. EMBO J. 2001 Aug 1;20(15):3910-6. PMID:11483494 doi:http://dx.doi.org/10.1093/emboj/20.15.3910
- ↑ Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein. EMBO J. 2001 Aug 1;20(15):3910-6. PMID:11483494 doi:http://dx.doi.org/10.1093/emboj/20.15.3910
- ↑ Tu Y, Kreinbring CA, Hill M, Liu C, Petsko GA, McCune CD, Berkowitz DB, Liu D, Ringe D. Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time. Biochemistry. 2018 Apr 13. doi: 10.1021/acs.biochem.8b00092. PMID:29630349 doi:http://dx.doi.org/10.1021/acs.biochem.8b00092
- ↑ Jhee KH, Kruger WD. The role of cystathionine beta-synthase in homocysteine metabolism. Antioxid Redox Signal. 2005 May-Jun;7(5-6):813-22. doi: 10.1089/ars.2005.7.813. PMID:15890029 doi:http://dx.doi.org/10.1089/ars.2005.7.813
- ↑ Tu Y, Kreinbring CA, Hill M, Liu C, Petsko GA, McCune CD, Berkowitz DB, Liu D, Ringe D. Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time. Biochemistry. 2018 Apr 13. doi: 10.1021/acs.biochem.8b00092. PMID:29630349 doi:http://dx.doi.org/10.1021/acs.biochem.8b00092
- ↑ Meier M, Oliveriusova J, Kraus JP, Burkhard P. Structural insights into mutations of cystathionine beta-synthase. Biochim Biophys Acta. 2003 Apr 11;1647(1-2):206-13. PMID:12686134
- ↑ Kraus JP, Janosik M, Kozich V, Mandell R, Shih V, Sperandeo MP, Sebastio G, de Franchis R, Andria G, Kluijtmans LA, Blom H, Boers GH, Gordon RB, Kamoun P, Tsai MY, Kruger WD, Koch HG, Ohura T, Gaustadnes M. Cystathionine beta-synthase mutations in homocystinuria. Hum Mutat. 1999;13(5):362-75. PMID:10338090 doi:<362::AID-HUMU4>3.0.CO;2-K http://dx.doi.org/10.1002/(SICI)1098-1004(1999)13:5<362::AID-HUMU4>3.0.CO;2-K
- ↑ Kozich V, Sokolova J, Klatovska V, Krijt J, Janosik M, Jelinek K, Kraus JP. Cystathionine beta-synthase mutations: effect of mutation topology on folding and activity. Hum Mutat. 2010 Jul;31(7):809-19. doi: 10.1002/humu.21273. PMID:20506325 doi:http://dx.doi.org/10.1002/humu.21273
- ↑ Meier M, Oliveriusova J, Kraus JP, Burkhard P. Structural insights into mutations of cystathionine beta-synthase. Biochim Biophys Acta. 2003 Apr 11;1647(1-2):206-13. PMID:12686134
- ↑ Meier M, Oliveriusova J, Kraus JP, Burkhard P. Structural insights into mutations of cystathionine beta-synthase. Biochim Biophys Acta. 2003 Apr 11;1647(1-2):206-13. PMID:12686134
- ↑ Kozich V, Sokolova J, Klatovska V, Krijt J, Janosik M, Jelinek K, Kraus JP. Cystathionine beta-synthase mutations: effect of mutation topology on folding and activity. Hum Mutat. 2010 Jul;31(7):809-19. doi: 10.1002/humu.21273. PMID:20506325 doi:http://dx.doi.org/10.1002/humu.21273
- ↑ Sperandeo MP, Candito M, Sebastio G, Rolland MO, Turc-Carel C, Giudicelli H, Dellamonica P, Andria G. Homocysteine response to methionine challenge in four obligate heterozygotes for homocystinuria and relationship with cystathionine beta-synthase mutations. J Inherit Metab Dis. 1996;19(3):351-6. PMID:8803779
- ↑ Mendes MI, Colaco HG, Smith DE, Ramos RJ, Pop A, van Dooren SJ, Tavares de Almeida I, Kluijtmans LA, Janssen MC, Rivera I, Salomons GS, Leandro P, Blom HJ. Reduced response of Cystathionine Beta-Synthase (CBS) to S-Adenosylmethionine (SAM): Identification and functional analysis of CBS gene mutations in Homocystinuria patients. J Inherit Metab Dis. 2014 Mar;37(2):245-54. doi: 10.1007/s10545-013-9647-6. Epub , 2013 Aug 23. PMID:23974653 doi:http://dx.doi.org/10.1007/s10545-013-9647-6
- ↑ Cui X, Navneet S, Wang J, Roon P, Chen W, Xian M, Smith SB. Analysis of MTHFR, CBS, Glutathione, Taurine, and Hydrogen Sulfide Levels in Retinas of Hyperhomocysteinemic Mice. Invest Ophthalmol Vis Sci. 2017 Apr 1;58(4):1954-1963. doi:, 10.1167/iovs.16-21247. PMID:28384716 doi:http://dx.doi.org/10.1167/iovs.16-21247
- ↑ Ansari R, Mahta A, Mallack E, Luo JJ. Hyperhomocysteinemia and neurologic disorders: a review. J Clin Neurol. 2014 Oct;10(4):281-8. doi: 10.3988/jcn.2014.10.4.281. Epub 2014, Oct 6. PMID:25324876 doi:http://dx.doi.org/10.3988/jcn.2014.10.4.281
- ↑ Miles EW, Kraus JP. Cystathionine beta-synthase: structure, function, regulation, and location of homocystinuria-causing mutations. J Biol Chem. 2004 Jul 16;279(29):29871-4. Epub 2004 Apr 15. PMID:15087459 doi:http://dx.doi.org/10.1074/jbc.R400005200
- ↑ Meier M, Oliveriusova J, Kraus JP, Burkhard P. Structural insights into mutations of cystathionine beta-synthase. Biochim Biophys Acta. 2003 Apr 11;1647(1-2):206-13. PMID:12686134
- ↑ Jhee KH, Kruger WD. The role of cystathionine beta-synthase in homocysteine metabolism. Antioxid Redox Signal. 2005 May-Jun;7(5-6):813-22. doi: 10.1089/ars.2005.7.813. PMID:15890029 doi:http://dx.doi.org/10.1089/ars.2005.7.813