We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1095
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
== History == | == History == | ||
=== Discovery of angiotensin receptors=== | === Discovery of angiotensin receptors=== | ||
| - | Researchers | + | Researchers had suspicions since the 1970s about the existence of different angiotensin receptors. However, tools to identify those distinct trans-membrane receptors became available only a decade later. Receptors binding assays identified angiotensin receptors in vitro using radioactive angiotensin. Results showed several types of angiotensin receptors, found in different tissues. The main receptors are AT1 and [https://en.wikipedia.org/wiki/Angiotensin_II_receptor_type_2 AT2] <ref>PMID:10821350</ref>. |
=== Nomenclature === | === Nomenclature === | ||
| Line 20: | Line 20: | ||
=== Recent studies === | === Recent studies === | ||
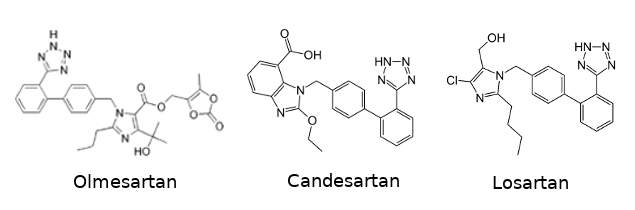
| - | + | Around 2015, researchers found the crystal structure of the receptor in complex with its antagonist [https://pubchem.ncbi.nlm.nih.gov/compound/ZD-7155-hydrochloride ZD7155] and with an inverse agonist [https://en.wikipedia.org/wiki/Olmesartan olmesartan]<ref> PMID: 26420482 </ref>. [https://en.wikipedia.org/wiki/X-ray_crystallography X-ray cryogenic-crystallography] has been used. They found similar conformation of the receptor when it is linked to the antagonist or to the inverse agonist. They have also found conserved molecular recognition modes. To complete this, they have performed mutagenesis experiments and managed to identify several residues in interaction with the ligand. | |
| - | The structure of this protein | + | The structure of this protein was solved in 2017 using another method called serial femtosecond crystallography, corresponding to the structure [http://proteopedia.org/wiki/index.php/4yay 4YAY] <ref name="Zhang2017">PMID:25913193</ref>. |
== Structure (function relationship) == | == Structure (function relationship) == | ||
=== Primary and secondary structure === | === Primary and secondary structure === | ||
| - | AT1 receptor consists | + | AT1 receptor consists of a 376 amino acid string <ref> [http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=4zud&template=main.html Protein Database (PDBsum): 4zud. European Bioinformatics (EBI); 2013.]</ref>. The protein is composed of |
<scene name='82/829348/Helix_a/1'>18 α helix</scene> | <scene name='82/829348/Helix_a/1'>18 α helix</scene> | ||
| - | and <scene name='82/829348/B_sheet/1'>3 β sheets</scene>. Moreover, 7 α | + | and <scene name='82/829348/B_sheet/1'>3 β sheets</scene>. Moreover, 7 α helixes are made of a majority of hydrophobic amino acids. These helixes are long enough to cross the membrane and create an <scene name='82/829348/Transmambrane_protein/1'>hydrophobic domain</scene> which is situated into the membrane. The human angiotensin receptor is therefore an α helical trans-membrane protein. |
| - | Since the angiotensin receptor belongs to the GPCRs family, those 7 α | + | Since the angiotensin receptor belongs to the GPCRs family, those 7 α helixes contain 3 extracellular and 3 intracellular loops. The N terminus corresponds to the extracellular domain. The C terminal domain is located intracellularly. |
=== Ligand binding pocket === | === Ligand binding pocket === | ||
Revision as of 16:46, 16 January 2020
| This Sandbox is Reserved from 25/11/2019, through 30/9/2020 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1091 through Sandbox Reserved 1115. |
To get started:
More help: Help:Editing |
Human Angiotensin Receptor
Angiotensin receptors of type 1 belong to the G protein coupled receptor (GPCR) family. These transmembrane proteins interact with angiotensin II, their ligand, and play a crucial role in the renin-angiotensin-aldosterone system. AT1 receptors are predominantly expressed in cardiovascular tissues including heart, endothelium, kidney, vascular smooth muscle cells as well as lungs, brain and adrenal cortex. [1] They are therefore important for the cardiovascular physiology.
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Thomas WG, Mendelsohn FA. Angiotensin receptors: form and function and distribution. Int J Biochem Cell Biol. 2003 Jun;35(6):774-9. doi:, 10.1016/s1357-2725(02)00263-7. PMID:12676163 doi:http://dx.doi.org/10.1016/s1357-2725(02)00263-7
- ↑ Kawai T, Forrester SJ, O'Brien S, Baggett A, Rizzo V, Eguchi S. AT1 receptor signaling pathways in the cardiovascular system. Pharmacol Res. 2017 Nov;125(Pt A):4-13. doi: 10.1016/j.phrs.2017.05.008. Epub, 2017 May 17. PMID:28527699 doi:http://dx.doi.org/10.1016/j.phrs.2017.05.008
- ↑ Goodfriend TL. Angiotensin receptors: history and mysteries. Am J Hypertens. 2000 Apr;13(4 Pt 1):442-9. doi: 10.1016/s0895-7061(99)00212-5. PMID:10821350 doi:http://dx.doi.org/10.1016/s0895-7061(99)00212-5
- ↑ Bumpus FM, Catt KJ, Chiu AT, DeGasparo M, Goodfriend T, Husain A, Peach MJ, Taylor DG Jr, Timmermans PB. Nomenclature for angiotensin receptors. A report of the Nomenclature Committee of the Council for High Blood Pressure Research. Hypertension. 1991 May;17(5):720-1. doi: 10.1161/01.hyp.17.5.720. PMID:2022414 doi:http://dx.doi.org/10.1161/01.hyp.17.5.720
- ↑ Zhang H, Unal H, Desnoyer R, Han GW, Patel N, Katritch V, Karnik SS, Cherezov V, Stevens RC. Structural Basis for Ligand Recognition and Functional Selectivity at Angiotensin Receptor. J Biol Chem. 2015 Sep 29. pii: jbc.M115.689000. PMID:26420482 doi:http://dx.doi.org/10.1074/jbc.M115.689000
- ↑ Zhang H, Unal H, Gati C, Han GW, Liu W, Zatsepin NA, James D, Wang D, Nelson G, Weierstall U, Sawaya MR, Xu Q, Messerschmidt M, Williams GJ, Boutet S, Yefanov OM, White TA, Wang C, Ishchenko A, Tirupula KC, Desnoyer R, Coe J, Conrad CE, Fromme P, Stevens RC, Katritch V, Karnik SS, Cherezov V. Structure of the Angiotensin receptor revealed by serial femtosecond crystallography. Cell. 2015 May 7;161(4):833-44. doi: 10.1016/j.cell.2015.04.011. Epub 2015 Apr, 23. PMID:25913193 doi:http://dx.doi.org/10.1016/j.cell.2015.04.011
- ↑ Protein Database (PDBsum): 4zud. European Bioinformatics (EBI); 2013.
- ↑ Fillion D, Cabana J, Guillemette G, Leduc R, Lavigne P, Escher E. Structure of the human angiotensin II type 1 (AT1) receptor bound to angiotensin II from multiple chemoselective photoprobe contacts reveals a unique peptide binding mode. J Biol Chem. 2013 Mar 22;288(12):8187-97. doi: 10.1074/jbc.M112.442053. Epub 2013, Feb 5. PMID:23386604 doi:http://dx.doi.org/10.1074/jbc.M112.442053
- ↑ Singh KD, Unal H, Desnoyer R, Karnik SS. Mechanism of Hormone Peptide Activation of a GPCR: Angiotensin II Activated State of AT1R Initiated by van der Waals Attraction. J Chem Inf Model. 2019 Jan 28;59(1):373-385. doi: 10.1021/acs.jcim.8b00583. Epub , 2019 Jan 16. PMID:30608150 doi:http://dx.doi.org/10.1021/acs.jcim.8b00583
- ↑ 10.0 10.1 Takezako T, Unal H, Karnik SS, Node K. Current topics in angiotensin II type 1 receptor research: Focus on inverse agonism, receptor dimerization and biased agonism. Pharmacol Res. 2017 Sep;123:40-50. doi: 10.1016/j.phrs.2017.06.013. Epub 2017 Jun, 23. PMID:28648738 doi:http://dx.doi.org/10.1016/j.phrs.2017.06.013