We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1102
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
Zika is a [https://en.wikipedia.org/wiki/Double-stranded_RNA_viruses double-stranded RNA virus]. The [https://en.wikipedia.org/wiki/Viral_replication genome replication] of double-stranded RNA viruses needs the intervention of a [https://en.wikipedia.org/wiki/Helicase helicase]. | Zika is a [https://en.wikipedia.org/wiki/Double-stranded_RNA_viruses double-stranded RNA virus]. The [https://en.wikipedia.org/wiki/Viral_replication genome replication] of double-stranded RNA viruses needs the intervention of a [https://en.wikipedia.org/wiki/Helicase helicase]. | ||
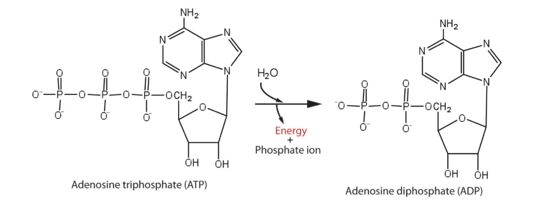
| - | Protein from Zika is composed of a protease domain at its [https://en.wikipedia.org/wiki/N-terminus N terminus] and a helicase domain at its [https://en.wikipedia.org/wiki/C-terminus C terminus]. The multifunctional helicase belongs to the superfamily 2 (SF2) helicase family. It is this helicase domain that has 5'-triphosphatase activity and that performs the critical and indispensable function of unwinding double-stranded RNA during | + | Protein from Zika is composed of a protease domain at its [https://en.wikipedia.org/wiki/N-terminus N terminus] and a helicase domain at its [https://en.wikipedia.org/wiki/C-terminus C terminus]. The multifunctional helicase belongs to the superfamily 2 (SF2) helicase family. It is this helicase domain that has 5'-triphosphatase activity and that performs the critical and indispensable function of unwinding double-stranded RNA during the step of replication. So it is an essential enzyme involved in the cycles of [https://en.wikipedia.org/wiki/ATP_hydrolysis ATP hydrolysis] and behaves as a key molecule in the RNA unwinding. Helicase performs ATP hydrolysis at the 5′ end of RNA to generate energy. During the hydrolysis, the phosphorus atom of the γ-phosphate group of the ATP molecule is cleaved by a water molecule; then, the γ-phosphate group is released with ADP. The break of the this high energy bound from ATP hydrolysis give enough energy to allow move the helicase along nucleic acid strands and unwind the double-stranded RNA genome. |
[[Image:ATPhydrolysis.jpg | thumb | upright=3,5 | Energy from ATP hydrolysis]] | [[Image:ATPhydrolysis.jpg | thumb | upright=3,5 | Energy from ATP hydrolysis]] | ||
| - | This step is among the necessary steps when it comes to viral RNA replication. It | + | This step is among the necessary steps when it comes to viral RNA replication. It creates a favorable environment for polymerization of RNA by an RNA-dependent RNA polymerase and the methylation of RNA by methyltransferase, which is essential for the pathogen development on its host. Moreover, it has been noticed that the viral capacity to replicate may be modified by the ATPase activity and therefore this enzyme would also change the host innate immune response. |
| - | + | <ref>Xu S, Ci Y, Wang, Yang, Zhang L, Xu C, Qin C, Shi L. Zika virus NS3 is a canonical RNA helicase stimulated by NS5 RNA polymerase.Nucleic Acids Res. 2019 Sep 19;47(16):8693-8707. doi: 10.1093/nar/gkz650.https://www.ncbi.nlm.nih.gov/pubmed/31361901</ref> | |
| - | + | ||
| Line 22: | Line 21: | ||
5y6n is a 1 chain protein of 438 residues. | 5y6n is a 1 chain protein of 438 residues. | ||
There are 2 main activities: ATP binding and helicase activity. | There are 2 main activities: ATP binding and helicase activity. | ||
| - | Helicase from Zika virus with a specific domain : a [https://en.wikipedia.org/wiki/Serine_protease serine protease]. It is a specific class of protein that hydrolysis peptide bonds from proteins. | + | Helicase from Zika virus with a specific domain: a [https://en.wikipedia.org/wiki/Serine_protease serine protease]. It is a specific class of protein that hydrolysis peptide bonds from proteins. |
| - | The helicase active site has a serine residue that serves as a nucleophilic amino acid. This residue is essential for the catalytic activity of the enzyme.The serine protases present a catalytic triad composed of 3 amino acids His57 Asp102 and Ser195<ref>Enrico Di Cera, Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, Box 8231, St. Louis, MO, USA; https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2675663/</ref>.These 3 amino acids have a specific interaction which is due to the folding of the protein. The tridimentional conformation of the protein brings closer these 3 residues. Their side chains have a specific function : | + | The helicase active site has a serine residue that serves as a nucleophilic amino acid. This residue is essential for the catalytic activity of the enzyme. The serine protases present a catalytic triad composed of 3 amino acids His57 Asp102 and Ser195<ref>Enrico Di Cera, Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, Box 8231, St. Louis, MO, USA; https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2675663/</ref>.These 3 amino acids have a specific interaction which is due to the folding of the protein. The tridimentional conformation of the protein brings closer these 3 residues. Their side chains have a specific function: |
[[Image:Serine protease catalysis.png | thumb | upright=3 | Serine protease catalysis]] | [[Image:Serine protease catalysis.png | thumb | upright=3 | Serine protease catalysis]] | ||
| - | '''Ser195''': the hydroxyl group plays the role of a nucleophile that | + | '''Ser195''': the hydroxyl group plays the role of a nucleophile that attacks the substrate. |
'''His57''': the nitrogen receives an hydrogen from the hydroxyl group of Ser195. | '''His57''': the nitrogen receives an hydrogen from the hydroxyl group of Ser195. | ||
| Line 32: | Line 31: | ||
'''Asp102''': this amino acid allows to increase the electronegativity of His157 nitrogen by making hydrogen bond with His157. | '''Asp102''': this amino acid allows to increase the electronegativity of His157 nitrogen by making hydrogen bond with His157. | ||
| - | All these reactions | + | All these reactions allow to cleave the substrate by attacking its carbonyl group. |
There are two ligands for zika helicase : ADP and Manganese II ion | There are two ligands for zika helicase : ADP and Manganese II ion | ||
| Line 38: | Line 37: | ||
-<scene name='82/829355/Mn/3'>Manganese II ion</scene> | -<scene name='82/829355/Mn/3'>Manganese II ion</scene> | ||
| - | The interactions between Zika helicase and these ligands are still studied to better understand their function and develop therapy. | + | There is a strong interrelationship between both ligands. The interactions between Zika helicase and these ligands are still studied to better understand their function and develop therapy. |
== Disease == | == Disease == | ||
Revision as of 17:21, 17 January 2020
| This Sandbox is Reserved from 25/11/2019, through 30/9/2020 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1091 through Sandbox Reserved 1115. |
To get started:
More help: Help:Editing |
5y6n- Zika virus helicase in complex with ADP
5y6n is a 1 chain protein structure. It’s the only helicase that belongs to zika virus. Zika helicase plays an important role in the pathogenocity of this virus.
| |||||||||||