Histone Lysine Methyltransferase SET7/9
From Proteopedia
(Difference between revisions)
| Line 26: | Line 26: | ||
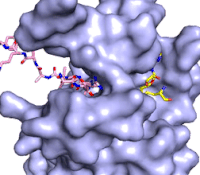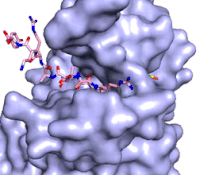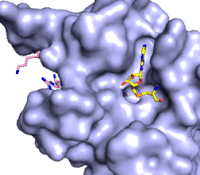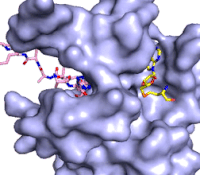
[[Image:KMTfinalmov2.gif|850 px|left|thumb|Figure 3: Gif-image of histone substrate (pink sticks) bound on one side of SET7/9 with the lysine target in the active site channel approaching S-adenosyl homocysteine (yellow sticks) bound on the opposite surface. The video was made using PyMOL, then converted to gif format using EZgif.]]The active site and binding pocket of KMT contain residues that optimize catalytic function and stability. First, the lysine of the substrate histone enters the active site via the <scene name='83/833386/Tyrosine_channel_2/3'>lysine access channel</scene> comprised of Tyr335 and Tyr337. The substrate lysine can be seen reaching through this channel to the SAM co-factor which is bound on the opposite face of the SET domain (Figure 3). With the histone substrate bound, the alkyl part of the lysine sidechain is stabilized by tyrosine residues in the <scene name='83/833386/Hydrophobic_packing/4'>substrate binding pocket</scene>, and polar residues are stabilized by hydrogen bonding interactions on the surface. Of note are the several interactions between SET7/9 and the Arg2 of the histone H3 peptide, including a hydrophobic interaction between Trp260 and the alkyl groups of the Arg2 side chain, and hydrogen bonds between Asp256 and Arg258 and Arg2 of the substrate. The <scene name='83/833386/Active_site_w_water/2'>active site</scene> itself contains the cofactor S-adenosyl methionine (SAM) which donates the methyl group in the reaction.<ref name="Xiao" /> In the active site scene, the structures depict the post-reaction result, where the Lys has been methylated and SAM has been converted to S-adenosyl homocysteine (SAH). | [[Image:KMTfinalmov2.gif|850 px|left|thumb|Figure 3: Gif-image of histone substrate (pink sticks) bound on one side of SET7/9 with the lysine target in the active site channel approaching S-adenosyl homocysteine (yellow sticks) bound on the opposite surface. The video was made using PyMOL, then converted to gif format using EZgif.]]The active site and binding pocket of KMT contain residues that optimize catalytic function and stability. First, the lysine of the substrate histone enters the active site via the <scene name='83/833386/Tyrosine_channel_2/3'>lysine access channel</scene> comprised of Tyr335 and Tyr337. The substrate lysine can be seen reaching through this channel to the SAM co-factor which is bound on the opposite face of the SET domain (Figure 3). With the histone substrate bound, the alkyl part of the lysine sidechain is stabilized by tyrosine residues in the <scene name='83/833386/Hydrophobic_packing/4'>substrate binding pocket</scene>, and polar residues are stabilized by hydrogen bonding interactions on the surface. Of note are the several interactions between SET7/9 and the Arg2 of the histone H3 peptide, including a hydrophobic interaction between Trp260 and the alkyl groups of the Arg2 side chain, and hydrogen bonds between Asp256 and Arg258 and Arg2 of the substrate. The <scene name='83/833386/Active_site_w_water/2'>active site</scene> itself contains the cofactor S-adenosyl methionine (SAM) which donates the methyl group in the reaction.<ref name="Xiao" /> In the active site scene, the structures depict the post-reaction result, where the Lys has been methylated and SAM has been converted to S-adenosyl homocysteine (SAH). | ||
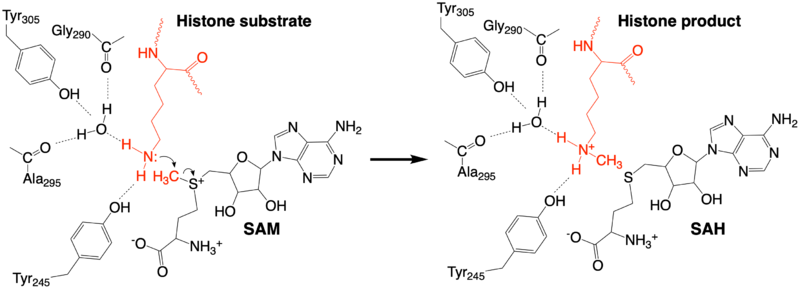
| - | The methylation reaction (Figure 4) is activated by several hydrogen bond interactions between active site residues and the substrate lysine. Specifically, the Tyr305 hydroxyl and the mainchain carbonyl oxygen of both Ala295 and Ser290 coordinate a buried water molecule that in turn coordinates a hydrogen of the lysine substrate. Additionally, the hydroxyl of Tyr245 also hydrogen bonds to the lysine substrate. These interactions enhance the nucleophilic nature of the amine nitrogen so that subsequent attack of the SAM methyl group carbon becomes favorable. Attack is further facilitated as the donor methyl is bound to a positively charged sulfur which will make a good leaving group when demthylated. Once the methyl group is transferred to the amine, the charge on the sulfur is resolved and thus SAM is converted to SAH.<ref name="Xiao" /> | + | The methylation reaction (Figure 4) is activated by several hydrogen bond interactions between active site residues and the substrate lysine. Specifically, the Tyr305 hydroxyl and the mainchain carbonyl oxygen of both Ala295 and Ser290 coordinate a buried water molecule that in turn coordinates a hydrogen of the lysine substrate. Additionally, the hydroxyl of Tyr245 also hydrogen bonds to the lysine substrate. These interactions enhance the nucleophilic nature of the amine nitrogen so that subsequent attack of the SAM methyl group carbon becomes favorable. Attack is further facilitated as the donor methyl is bound to a positively charged sulfur which will make a good leaving group when demthylated. Once the methyl group is transferred to the amine, the charge on the sulfur is resolved and thus SAM is converted to SAH.<ref name="Xiao" /> |
===The C-Terminal Domain=== | ===The C-Terminal Domain=== | ||
| - | The C-terminal domain of lysine methyltransferase consists of a beta-hairpin and alpha helix that serve as a 'cap' for the SET domain. The overall structure of the <scene name='83/833386/C_terminal_domain/1'>C-terminal domain (residues 340-366)</scene> provides various interactions that facilitate binding of substrate to the SET domain (residues 193-344).<ref name="Xiao" /> Hydrophobic interactions between the C-terminal domain and the SET domain are mainly responsible in forming the access channel for the substrate and assist in deprotonation of the lysine. Residues 337-349 create a pro-gly rich <scene name='83/833386/Beta-hairpin/5'>beta-hairpin</scene> that stabilizes the orientation of two tyrosine residues, Tyr 335 and Tyr337, that form the lysine access channel. Furthermore, there is substantial hydrophobic packing of the C-terminal helix against the SET domain using <scene name='83/833386/C_terminal_domain/4'>residues Phe299, Tyr353, Leu357 and Phe360 </scene>. These interactions position the indole ring of Trp352 in the C-terminal helix to <scene name='83/833386/C_terminal_domain/6'>stack with the π-cloud of the adenine base of the SAM co-factor</scene> as well as allowing Glu356 to hydrogen bond with N6 of the adenine ring.<ref name="Xiao" /> | + | The C-terminal domain of lysine methyltransferase consists of a beta-hairpin and alpha helix that serve as a 'cap' for the SET domain. The overall structure of the <scene name='83/833386/C_terminal_domain/1'>C-terminal domain (residues 340-366)</scene> provides various interactions that facilitate binding of substrate to the SET domain (residues 193-344).<ref name="Xiao" /> Hydrophobic interactions between the C-terminal domain and the SET domain are mainly responsible in forming the access channel for the substrate and assist in deprotonation of the lysine. Residues 337-349 create a pro-gly rich <scene name='83/833386/Beta-hairpin/5'>beta-hairpin</scene> that stabilizes the orientation of two tyrosine residues, Tyr 335 and Tyr337, that form the lysine access channel. Furthermore, there is substantial hydrophobic packing of the C-terminal helix against the SET domain using <scene name='83/833386/C_terminal_domain/4'>residues Phe299, Tyr353, Leu357 and Phe360 </scene>. These interactions position the indole ring of Trp352 in the C-terminal helix to <scene name='83/833386/C_terminal_domain/6'>stack with the π-cloud of the adenine base of the SAM co-factor</scene> as well as allowing Glu356 to hydrogen bond with N6 of the adenine ring.<ref name="Xiao" />[[Image:KMT_mechanism1.png|800px|center|thumb|Figure 4: The proposed KMT Mechanism]] |
Revision as of 13:45, 23 January 2020
Histone Lysine Methyltransferase, SET7/9: A transcriptional activator
| |||||||||||
Student Contributors
Lauryn Padgett, Alexandra Pentala, Madeleine Wilson
Proteopedia Page Contributors and Editors (what is this?)
Mark Macbeth, Michal Harel, Valentine J Klimkowski, Angel Herraez