Histone acetyltransferase 1-2 Complex (HAT1/2)
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
<StructureSection load='4PSW' size='350' frame='true' side='right' caption='HAT1-HAT2 Complex pdb: 4PSW' scene='83/834210/Overview/2'> | <StructureSection load='4PSW' size='350' frame='true' side='right' caption='HAT1-HAT2 Complex pdb: 4PSW' scene='83/834210/Overview/2'> | ||
=Histones= | =Histones= | ||
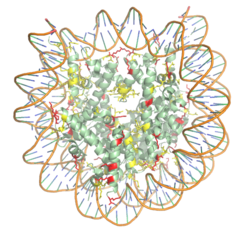
| - | [https://en.wikipedia.org/wiki/Histone Histones] are proteins found in the cell nucleus that are the key building blocks of [https://en.wikipedia.org/wiki/Chromatin chromatin] and are essential for proper DNA packaging and [https://en.wikipedia.org/wiki/Transcription_(biology) transcription]. In the first step of [https://www.hhmi.org/biointeractive/how-dna-packaged DNA packaging], two copies of the four core histone proteins ([https://en.wikipedia.org/wiki/Histone_H1 H1], [https://en.wikipedia.org/wiki/Histone_H2A H2A], [https://en.wikipedia.org/wiki/Histone_H3 H3], and [https://en.wikipedia.org/wiki/Histone_H4 H4]) form an [https://en.wikipedia.org/wiki/Histone_octamer octamer] that DNA wraps around, forming the [https://en.wikipedia.org/wiki/Nucleosome nucleosome]. 20-24% of residues making up the histone octamer are arginine and lysine, causing a net positive charge, especially at the outer surfaces of the histone core where negatively charged DNA is bound (Figure 1). <ref> Watson, J D, et al. Molecular Biology of the Gene (Seventh Edition). (2014) Boston, MA: Benjamin-Cummings Publishing Company. </ref><ref name="Watanabe"> PMID: 20100606 </ref> The positively charged residues of the histone core tails are often subject to chemical modifications | + | [https://en.wikipedia.org/wiki/Histone Histones] are proteins found in the cell nucleus that are the key building blocks of [https://en.wikipedia.org/wiki/Chromatin chromatin] and are essential for proper DNA packaging and the regulation of [https://en.wikipedia.org/wiki/Transcription_(biology) transcription]. In the first step of [https://www.hhmi.org/biointeractive/how-dna-packaged DNA packaging], two copies of the four core histone proteins ([https://en.wikipedia.org/wiki/Histone_H1 H1], [https://en.wikipedia.org/wiki/Histone_H2A H2A], [https://en.wikipedia.org/wiki/Histone_H3 H3], and [https://en.wikipedia.org/wiki/Histone_H4 H4]) form an [https://en.wikipedia.org/wiki/Histone_octamer octamer] that DNA wraps around, forming the [https://en.wikipedia.org/wiki/Nucleosome nucleosome]. 20-24% of residues making up the histone octamer are arginine and lysine, causing a net positive charge, especially at the outer surfaces of the histone core where negatively charged DNA is bound (Figure 1). <ref> Watson, J D, et al. Molecular Biology of the Gene (Seventh Edition). (2014) Boston, MA: Benjamin-Cummings Publishing Company. </ref><ref name="Watanabe"> PMID: 20100606 </ref> The positively charged residues of the histone core tails are often subject to chemical modifications that can regulate the processes of DNA repair, replication, transcription, and heterochromatin maintenance. |
[[Image:Histone_NEWEST_w_DNA.png|250 px|right|thumb|Figure 1. The nucleosome structure consisting of the histone octamer and DNA. Arginine residues are shown in yellow, lysine residues are shown in red. PDB: 3kwq]] | [[Image:Histone_NEWEST_w_DNA.png|250 px|right|thumb|Figure 1. The nucleosome structure consisting of the histone octamer and DNA. Arginine residues are shown in yellow, lysine residues are shown in red. PDB: 3kwq]] | ||
=Histone Modification= | =Histone Modification= | ||
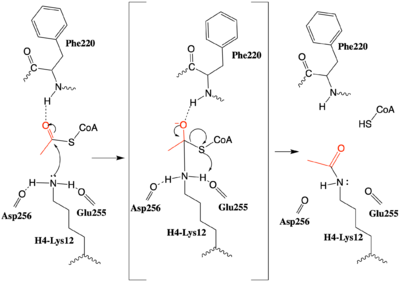
| - | Histones can be reversibly modified in a variety of ways, including: methylation, acetylation, phosphorylation, and ubiquitination. These modifications all result in either the condensation or relaxation of DNA and consequently repressing or activating transcription. Histone acetylation is a histone modification that involves the transfer of an acetyl group from the | + | Histones can be reversibly modified in a variety of ways, including: methylation, acetylation, phosphorylation, and ubiquitination. These modifications all result in either the condensation or relaxation of DNA and consequently repressing or activating transcription. Histone acetylation is a histone modification that involves the transfer of an acetyl group from the co-factor Acetyl Coenzyme A ([https://en.wikipedia.org/wiki/Acetyl-CoA acetyl-CoA]) to the ε-amino group of a lysine residue in a histone protein. This reaction is catalyzed by various histone acetyltransferase ([https://en.wikipedia.org/wiki/Histone_acetyltransferase HAT]) enzymes. Lysine acetylation is an important [https://en.wikipedia.org/wiki/Epigenetics epigenetic] marker as it removes the positive charge of the lysine, thus reducing the strength of the interaction between DNA and hisotones. The net effect of acetylation is the relaxation of the DNA into [http://www.example.com euchromatin] which is more transcriptionally active. |
=HAT1 Background = | =HAT1 Background = | ||
| - | HAT1 was the first of the enzymes to be identified in the HAT family | + | HAT1 catalyzes the acetylation of lysine residues and was the first of the enzymes to be identified in the HAT family. <ref name="Yang"> PMID: 24835250 </ref> Deletion of the HAT gene caused a loss of acetylation on H4K5 and H4K12, leading to the conclusion that HAT1 is the sole enzyme responsible for this conserved histone modification.<ref name="Parthun"> PMID: 8858151 </ref> The HAT2 enzyme was later identified as a binding partner for HAT1 and it modulates the substrate specificity of HAT1.<ref name="Yang"/> The HAT1/HAT2 complex is highly specific for H4K12 acetylation. |
= HAT1/HAT2 Complex Structure = | = HAT1/HAT2 Complex Structure = | ||
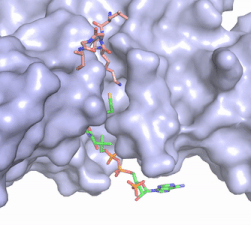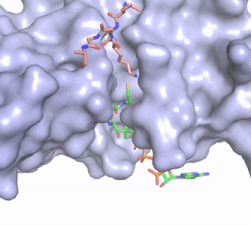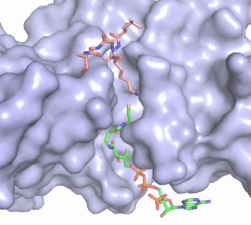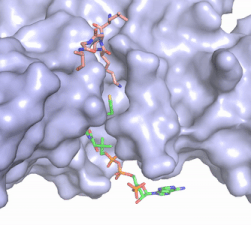
| - | The structure of the <scene name='83/834210/Overview/2'>HAT1-HAT2 complex</scene> was determined by X-ray diffraction at 2.0Å resolution. The complex was crystallized in the presence of coenzyme A ([https://en.wikipedia.org/wiki/Coenzyme_A CoA]) and a H4 N-terminal peptide (1-48). The complex has four components (HAT1, HAT2, H4 peptide, and CoA) seen with a 1:1:1:1 stoichiometry. While residues 1-48 of the yeast H4 protein were included in the crystallization, only residues 9-46 were well resolved in the refined structure. Similarly, residues 1-4 in the HAT1 subunit and residues 1-6, 87-104, and 391-401 in the HAT2 subunit were not resolved <ref name="Yang"/>. | + | The structure of the <scene name='83/834210/Overview/2'>HAT1-HAT2 complex</scene> from the yeast ''Saccharomyces cerevisiae'' was determined by X-ray diffraction at 2.0Å resolution. The complex was crystallized in the presence of coenzyme A ([https://en.wikipedia.org/wiki/Coenzyme_A CoA]) and a H4 N-terminal peptide (1-48). The complex has four components (HAT1, HAT2, H4 peptide, and CoA) seen with a 1:1:1:1 stoichiometry. While residues 1-48 of the yeast H4 protein were included in the crystallization, only residues 9-46 were well resolved in the refined structure. Similarly, residues 1-4 in the HAT1 subunit and residues 1-6, 87-104, and 391-401 in the HAT2 subunit were not resolved <ref name="Yang"/>. |
HAT1 is not catalytically active until it binds with HAT2 to form the complex. <ref name="Wu"> PMID: 22615379 </ref> The <scene name='83/834210/Hat1_-_chain_a/2'>HAT1 structure</scene> includes 317 residues and contains the binding site for acetyl-coenzyme A. <scene name='83/834210/Hat2_-_chain_b/2'>HAT2</scene> is 401 residues in a beta-propeller formation with C7 symmetry. Bound to the Hat1-Hat2 complex is a <scene name='83/834210/Histone_4/3'>peptide substrate</scene> comprised of residues 9-46 of the histone protein H4. | HAT1 is not catalytically active until it binds with HAT2 to form the complex. <ref name="Wu"> PMID: 22615379 </ref> The <scene name='83/834210/Hat1_-_chain_a/2'>HAT1 structure</scene> includes 317 residues and contains the binding site for acetyl-coenzyme A. <scene name='83/834210/Hat2_-_chain_b/2'>HAT2</scene> is 401 residues in a beta-propeller formation with C7 symmetry. Bound to the Hat1-Hat2 complex is a <scene name='83/834210/Histone_4/3'>peptide substrate</scene> comprised of residues 9-46 of the histone protein H4. | ||
Revision as of 21:28, 26 January 2020
Histone Acetyltransferase HAT1/HAT2 Complex, Saccharomyces cerevisiae
| |||||||||||
Student Contributors
Morgan Buckley, Jordan Finch, Caitlin Gaich, Kiran Kaur, Emily Leiderman, Ben Nick
Proteopedia Page Contributors and Editors (what is this?)
Mark Macbeth, Angel Herraez, Michal Harel, Valentine J Klimkowski