We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Ivan E. Wang/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
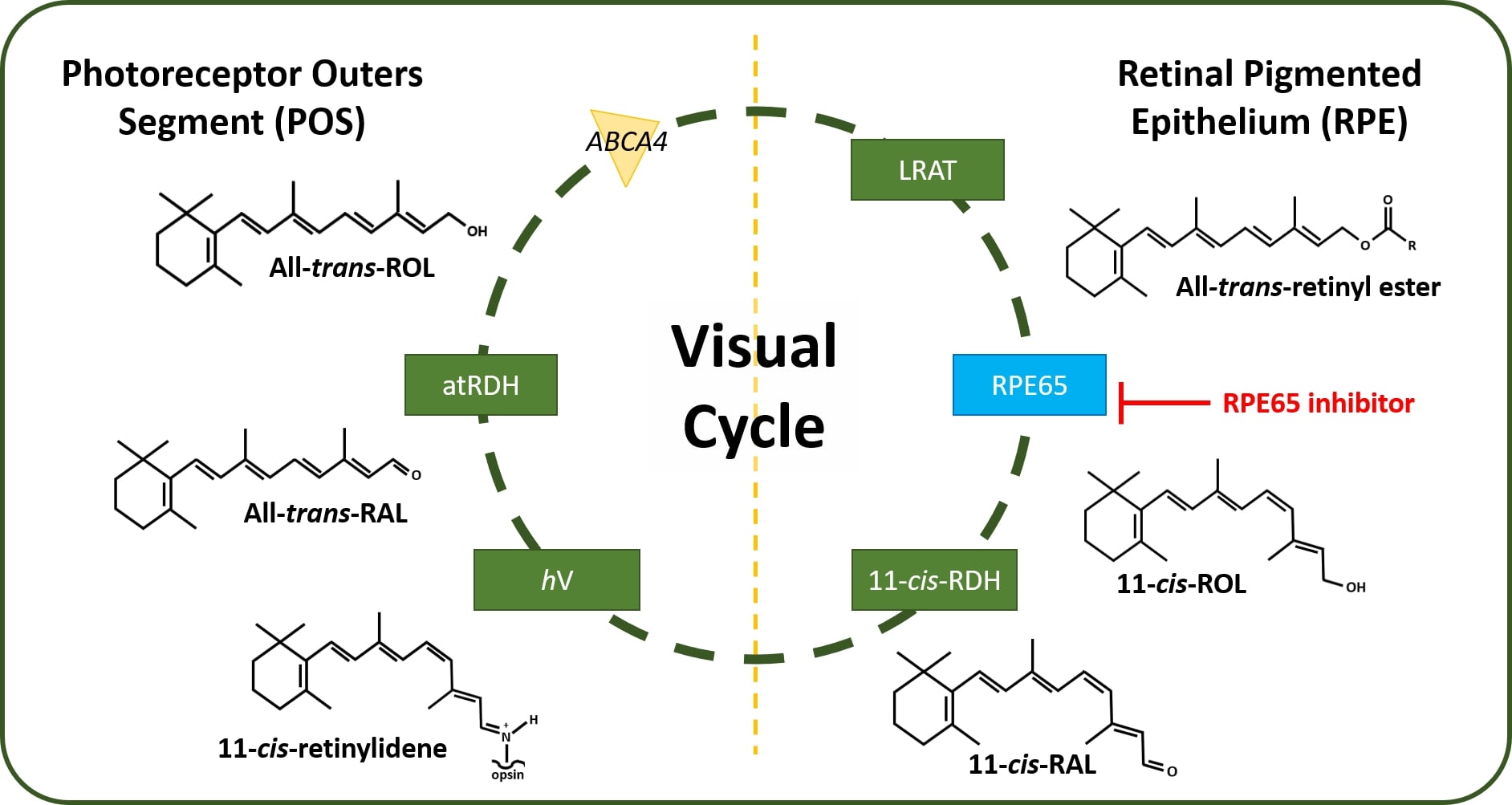
Retinal pigment epithelium 65 (RPE65) also known as retinoid isomerohydrolase is a 65 kDa enzyme located within the human retinal pigment epithelium cells responsible for the chemical conversion of ''all-trans''-retinyl ester to ''11-cis''-retinol in the visual cycle. | Retinal pigment epithelium 65 (RPE65) also known as retinoid isomerohydrolase is a 65 kDa enzyme located within the human retinal pigment epithelium cells responsible for the chemical conversion of ''all-trans''-retinyl ester to ''11-cis''-retinol in the visual cycle. | ||
</StructureSection> | </StructureSection> | ||
| - | |||
| - | (PLACEHOLDER This is a default text for your page '''Ivan E. Wang/Sandbox 1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
| - | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue.) | ||
= Function = | = Function = | ||
| Line 14: | Line 11: | ||
== Enzymatic Catalyzed Reaction == | == Enzymatic Catalyzed Reaction == | ||
(PLACEHOLDER) | (PLACEHOLDER) | ||
| - | [[Image:Visual Cycle.jpg | + | [[Image:Visual Cycle.jpg|left|alt=Figure 2: Human Visual Cycle| Figure 2: The canonical Visual Cycle in Humans <ref>DOI 10.1016/j.bbadis.2018.04.014</ref>]] |
Revision as of 23:00, 28 January 2020
Retinal Pigment Epithelium 65 (Retinoid Isomerohydrolase) complex with (R/S)-emixustat and palmitate
| |||||||||||
Contents |
Function
Structural Features of RPE65
(PLACEHOLDER)
Enzymatic Catalyzed Reaction
(PLACEHOLDER)
Protein-Ligand Interaction
Endogenous Ligand
(PLACEHOLDER)
Exogenous Ligand
| |||||||||||
| |||||||||||
R/S Enantiomers Differences
(PLACEHOLDER)
Disease Implications
(PLACEHOLDER)
Medical Relevance
(PLACEHOLDER)
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
References
- ↑ Shin Y, Moiseyev G, Petrukhin K, Cioffi CL, Muthuraman P, Takahashi Y, Ma JX. A novel RPE65 inhibitor CU239 suppresses visual cycle and prevents retinal degeneration. Biochim Biophys Acta Mol Basis Dis. 2018 Jul;1864(7):2420-2429. doi:, 10.1016/j.bbadis.2018.04.014. Epub 2018 Apr 21. PMID:29684583 doi:http://dx.doi.org/10.1016/j.bbadis.2018.04.014