User:Ivan E. Wang/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
= Introduction to the RPE65 = | = Introduction to the RPE65 = | ||
| - | |||
<StructureSection load='4rsc' size='500' side='right' caption='Figure 1: Emixustat and palmitate bound in the active site of RPE65' scene=''> | <StructureSection load='4rsc' size='500' side='right' caption='Figure 1: Emixustat and palmitate bound in the active site of RPE65' scene=''> | ||
| - | |||
Retinal pigment epithelium 65 (RPE65) also known as retinoid isomerohydrolase is a 65 kDa enzyme located within the human retinal pigment epithelium cells. PRE65 is responsible for the chemical conversion of ''all-trans''-retinyl ester to ''11-cis''-retinol in the visual cycle. | Retinal pigment epithelium 65 (RPE65) also known as retinoid isomerohydrolase is a 65 kDa enzyme located within the human retinal pigment epithelium cells. PRE65 is responsible for the chemical conversion of ''all-trans''-retinyl ester to ''11-cis''-retinol in the visual cycle. | ||
</StructureSection> | </StructureSection> | ||
| - | = | + | = Structure and Activity = |
| - | == Structural | + | == Structural Analysis of RPE65 == |
(PLACEHOLDER) | (PLACEHOLDER) | ||
| - | == Enzymatic | + | == Enzymatic Activity of RPE65 == |
(PLACEHOLDER) | (PLACEHOLDER) | ||
[[Image:Visual Cycle.jpg|thumb|right|512 px|alt=Figure 2: Human Visual Cycle| Figure 2: The canonical Visual Cycle in Humans <ref>DOI 10.1016/j.bbadis.2018.04.014</ref>]] | [[Image:Visual Cycle.jpg|thumb|right|512 px|alt=Figure 2: Human Visual Cycle| Figure 2: The canonical Visual Cycle in Humans <ref>DOI 10.1016/j.bbadis.2018.04.014</ref>]] | ||
| Line 21: | Line 19: | ||
== Exogenous Ligand == | == Exogenous Ligand == | ||
| + | === (''R'')-Emixustat (ACU-4429) === | ||
<StructureSection load='4ryy' size='340' side='right' caption='Figure 3:(R)-emixustat and palmitate bound in the active site of RPE65' scene=''> | <StructureSection load='4ryy' size='340' side='right' caption='Figure 3:(R)-emixustat and palmitate bound in the active site of RPE65' scene=''> | ||
| - | === (''R'')-Emixustat (ACU-4429) === | ||
| - | |||
(''R'')-emixustat (ACU-4429) is an investigational small molecule inhibitor of RPE65 first invented by a British-American chemist, Ian L. Scott. Formulated as an hydrochloride salt, (''R'')-emixustat hydrochloride is taken by mouth and functions as a visual cycle modulator (VCM) to reduce toxic retinal byproducts such as A2E. | (''R'')-emixustat (ACU-4429) is an investigational small molecule inhibitor of RPE65 first invented by a British-American chemist, Ian L. Scott. Formulated as an hydrochloride salt, (''R'')-emixustat hydrochloride is taken by mouth and functions as a visual cycle modulator (VCM) to reduce toxic retinal byproducts such as A2E. | ||
| - | |||
In 2008, Acucela Inc. partnered with Otsuka Pharmaceutical Company for the continued development of (''R'')-emixustat as a potential inhibitor of RPE65. Currently (''R'')-emixustat is in Phase III clinical trials in the United States for the potential treatment of Stargard's disease, a juvenile form of atrophic (dry) age dependent macular degeneration (AMD). Additionally, (''R'')-emixustat is investigated as potential therapy for diabetic retinopathy and diabetic macular edema. | In 2008, Acucela Inc. partnered with Otsuka Pharmaceutical Company for the continued development of (''R'')-emixustat as a potential inhibitor of RPE65. Currently (''R'')-emixustat is in Phase III clinical trials in the United States for the potential treatment of Stargard's disease, a juvenile form of atrophic (dry) age dependent macular degeneration (AMD). Additionally, (''R'')-emixustat is investigated as potential therapy for diabetic retinopathy and diabetic macular edema. | ||
| - | |||
</StructureSection> | </StructureSection> | ||
| + | === (''S'')-Emixustat === | ||
<StructureSection load='4ryz' size='340' side='right' caption='Figure 4:(S)-emixustat and palmitate bound in the active site of RPE65' scene=''> | <StructureSection load='4ryz' size='340' side='right' caption='Figure 4:(S)-emixustat and palmitate bound in the active site of RPE65' scene=''> | ||
| - | === (''S'')-Emixustat === | ||
(PLACEHOLDER) | (PLACEHOLDER) | ||
| - | |||
</StructureSection> | </StructureSection> | ||
| Line 44: | Line 38: | ||
= Medical Relevance = | = Medical Relevance = | ||
(PLACEHOLDER) | (PLACEHOLDER) | ||
| - | |||
| - | |||
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
Revision as of 23:23, 28 January 2020
Contents |
Introduction to the RPE65
| |||||||||||
Structure and Activity
Structural Analysis of RPE65
(PLACEHOLDER)
Enzymatic Activity of RPE65
(PLACEHOLDER)
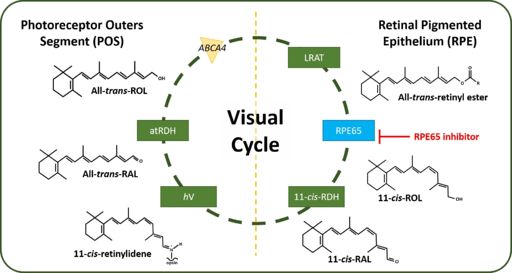
Figure 2: The canonical Visual Cycle in Humans [1]
Protein-Ligand Interaction
Endogenous Ligand
(PLACEHOLDER)
Exogenous Ligand
(R)-Emixustat (ACU-4429)
| |||||||||||
(S)-Emixustat
| |||||||||||
R/S Enantiomers Differences
(PLACEHOLDER)
Disease Implications
(PLACEHOLDER)
Medical Relevance
(PLACEHOLDER)
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
References
- ↑ Shin Y, Moiseyev G, Petrukhin K, Cioffi CL, Muthuraman P, Takahashi Y, Ma JX. A novel RPE65 inhibitor CU239 suppresses visual cycle and prevents retinal degeneration. Biochim Biophys Acta Mol Basis Dis. 2018 Jul;1864(7):2420-2429. doi:, 10.1016/j.bbadis.2018.04.014. Epub 2018 Apr 21. PMID:29684583 doi:http://dx.doi.org/10.1016/j.bbadis.2018.04.014
