Comparing serial X-ray crystallography and microcrystal electron diffraction (MicroED) as methods for routine structure determination from small macromolecular crystals
Alexander M Wolff, Iris D Young, Raymond G Sierra, Aaron S Brewster, Michael W Martynowycz, Eriko Nango, Michihiro Sugahara, Takanori Nakane, Kazutaka Ito, Andrew Aquila, Asmit Bhowmik, Justin T Biel, Sergio Carbajo, Aina E Cohen; Saul Cortez, Ana Gonzalez, Tomoya Hino, Dohyun Im, Jake D Koralek, Minoru Kubo, Tomas S Lazarou, Takashi Nomura, Shigeki Owada, Avi J. Samelson, Tomoyuki Tanaka, Rie Tanaka, Erin M Thompson, Henry van den Bedem, Rahel A Woldeyes, Fumiaki Yumoto, Wei Zhao, Kensuke Tono, Sébastien Boutet, So Iwata, Tamir Gonen, Nicholas K Sauter, James S Fraser, Michael C Thompson [1]
Molecular Tour
Crystallography capitalizes on repetition. Large crystals have more repeating units, which results in a stronger signal, enabling us to build a high-resolution model. When a crystal is too small, we cannot measure high-resolution data points, and our picture of the molecule is less crisp. Traditionally, we overcame this problem by growing larger crystals, but an alternative strategy is to get a brighter light source. X-ray lasers (XFELs) are the latest innovation on this front, and are one-billion times brighter than previous X-ray sources. In theory, this means we can solve structures from crystals one-billionth the size of those used for traditional experiments. Another alternative is to use electrons, which interact with molecules more strongly than X-rays. Due to the strength of the interaction between electrons and matter, we can use electron diffraction (microED) to solve structures from even smaller crystals.
With these recent additions to our toolbox, we wondered whether each method would provide the same view of the molecules within the crystal. One primary difference between the two methods is the temperature at which samples are prepared. For microED, samples are frozen using liquid nitrogen, whereas XFEL samples are prepared and investigated at room temperature. In the Fraser Lab, we have a longstanding interest in the effect of temperature upon protein structure. We have demonstrated that cryogenic freezing can alter the conformations a molecule adopts. Nonetheless, since we could use very small crystals for microED, we wondered whether we could freeze them quickly enough to lock molecules into their room-temperature conformations.
Our model system for this work was Cyclophilin A (CypA), which we had previously studied at multiple temperatures. As it turned out, both microED and XFELs enabled us to solve high-resolution structures of CypA, but the conformations of the protein were not the same. In the microED structure, CypA adopted a single conformation previously seen in cryogenic X-ray studies, whereas the XFEL data revealed multiple conformations, as seen in previous room-temperature X-ray studies. So, our work reveals that temperature is a defining difference between structures solved using XFELs and those solved using microED. For many proteins, different conformations are visible at cryogenic temperatures when compared to room temperature. This means that these two methods provide complementary views into the molecular landscape, in addition to enabling collection of high-resolution data from small crystals.
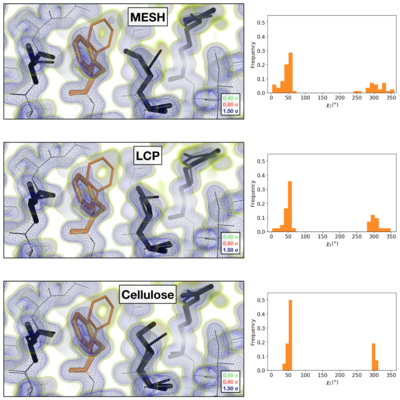
Comparison of the 2mFoFc map and the refined multi-conformer model produced from each serial XFEL experiment. Maps were visualized at multiple contour levels to show evidence for alternative conformations (see static image above). Following multi-conformer refinement, ensembles were generated from each model using phenix.ensemble_refine. In the right panel, a histogram of the Chi1 angles for residue 113 is plotted for the ensemble. Multi-conformer models plus maps, and the distribution of Chi1 angles across the ensemble models are similar for all three XFEL data sets:
References
- ↑ Wolff AM, Young ID, Sierra RG, Brewster AS, Martynowycz MW, Nango E, Sugahara M, Nakane T, Ito K, Aquila A, Bhowmick A, Biel JT, Carbajo S, Cohen AE, Cortez S, Gonzalez A, Hino T, Im D, Koralek JD, Kubo M, Lazarou TS, Nomura T, Owada S, Samelson AJ, Tanaka T, Tanaka R, Thompson EM, van den Bedem H, Woldeyes RA, Yumoto F, Zhao W, Tono K, Boutet S, Iwata S, Gonen T, Sauter NK, Fraser JS, Thompson MC. Comparing serial X-ray crystallography and microcrystal electron diffraction (MicroED) as methods for routine structure determination from small macromolecular crystals. IUCrJ. 2020 Feb 26;7(Pt 2):306-323. doi: 10.1107/S205225252000072X. eCollection, 2020 Mar 1. PMID:32148858 doi:http://dx.doi.org/10.1107/S205225252000072X