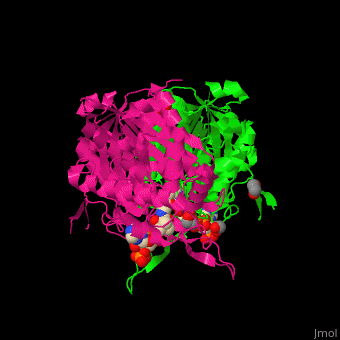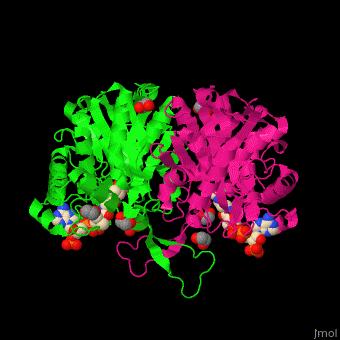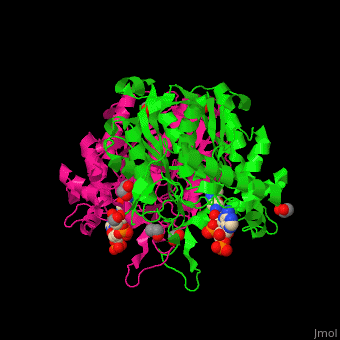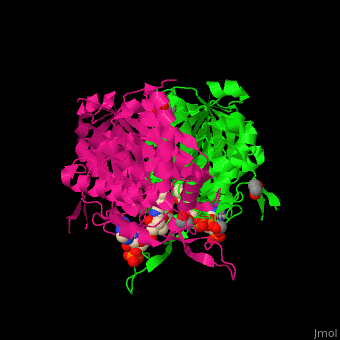We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Thiolase
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
== Structural highlights == | == Structural highlights == | ||
The <scene name='44/447919/Cv/8'>active site of 3-ketoacyl-CoA thiolase contains CoA</scene><ref>PMID:25478839</ref>. Water molecules are shown as red spheres. | The <scene name='44/447919/Cv/8'>active site of 3-ketoacyl-CoA thiolase contains CoA</scene><ref>PMID:25478839</ref>. Water molecules are shown as red spheres. | ||
| + | == 3D Structures of Thiolase == | ||
| + | [[Thiolase 3D structures]] | ||
| + | |||
</StructureSection> | </StructureSection> | ||
== 3D Structures of Thiolase == | == 3D Structures of Thiolase == | ||
| Line 19: | Line 22: | ||
*3-ketoacyl-CoA thiolase | *3-ketoacyl-CoA thiolase | ||
| - | **[[2wu9]], [[2c7y]], [[2c7z]] – KCT peroxisomal residues 36-462 – ''Arabidopsis thaliana''<br /> | ||
| - | **[[2wua]] - KCT peroxisomal residues 17-449 (mutant) – sunflower<br /> | ||
**[[2iik]] – hKCT peroxisomal – human<br /> | **[[2iik]] – hKCT peroxisomal – human<br /> | ||
**[[4c2k]] – hKCT mitochondrial <br /> | **[[4c2k]] – hKCT mitochondrial <br /> | ||
**[[4c2j]] – hKCT mitochondrial + CoA<br /> | **[[4c2j]] – hKCT mitochondrial + CoA<br /> | ||
| + | **[[2wu9]], [[2c7y]], [[2c7z]] – KCT peroxisomal residues 36-462 – ''Arabidopsis thaliana''<br /> | ||
| + | **[[2wua]] - KCT peroxisomal residues 17-449 (mutant) – sunflower<br /> | ||
| + | **[[1afw]], [[1pxt]] – yKCT peroxisomal – yeast<br /> | ||
**[[3goa]] – KCT – ''Salmonella typhimurium''<br /> | **[[3goa]] – KCT – ''Salmonella typhimurium''<br /> | ||
**[[5lp7]] – KCT – ''Bacillus subtilis'' <br /> | **[[5lp7]] – KCT – ''Bacillus subtilis'' <br /> | ||
| - | **[[1afw]], [[1pxt]] – yKCT peroxisomal – yeast<br /> | ||
**[[5cbq]] – MsKCT – ''Mycobacterium smegmatis'' <br /> | **[[5cbq]] – MsKCT – ''Mycobacterium smegmatis'' <br /> | ||
**[[4w61]] – ReKCT – ''Ralstonia eutropha''<br /> | **[[4w61]] – ReKCT – ''Ralstonia eutropha''<br /> | ||
| Line 43: | Line 46: | ||
*Acetoacetyl-CoA thiolase or acetyl-CoA acetyltransferase | *Acetoacetyl-CoA thiolase or acetyl-CoA acetyltransferase | ||
| - | **[[1m4s]], [[1m4t]], [[1dlu]], [[1qfl]] - ZrACT – ''Zoogloea ramigera''<br /> | ||
| - | **[[2wku]], [[2wl6]], [[1m1t]], [[1m3k]] - ZrACT (mutant) <br /> | ||
**[[2ib7]], [[2ib8]], [[2ib9]], [[2ibu]], [[2ibw]], [[2iby]], [[2f2s]] – hACT mitochondrial<br /> | **[[2ib7]], [[2ib8]], [[2ib9]], [[2ibu]], [[2ibw]], [[2iby]], [[2f2s]] – hACT mitochondrial<br /> | ||
**[[1wl5]] - hACT cytosolic<br /> | **[[1wl5]] - hACT cytosolic<br /> | ||
| + | **[[5xyj]], [[5xz5]] – yACT <br /> | ||
| + | **[[1m4s]], [[1m4t]], [[1dlu]], [[1qfl]] - ZrACT – ''Zoogloea ramigera''<br /> | ||
| + | **[[2wku]], [[2wl6]], [[1m1t]], [[1m3k]] - ZrACT (mutant) <br /> | ||
**[[4dd5]], [[4e1l]] – ACT – ''Clostridium difficile''<br /> | **[[4dd5]], [[4e1l]] – ACT – ''Clostridium difficile''<br /> | ||
**[[4xl2]], [[4xl3]], [[4n44]], [[4n45]] – CaACT – ''Clostridium acetobutylicum''<br /> | **[[4xl2]], [[4xl3]], [[4n44]], [[4n45]] – CaACT – ''Clostridium acetobutylicum''<br /> | ||
| Line 52: | Line 56: | ||
**[[5f0v]], [[5f38]], [[4wys]] – ACT – ''Escherichia coli'' <br /> | **[[5f0v]], [[5f38]], [[4wys]] – ACT – ''Escherichia coli'' <br /> | ||
**[[4o9a]], [[4o99]], [[4o9c]], [[4nzs]] – ReACT <br /> | **[[4o9a]], [[4o99]], [[4o9c]], [[4nzs]] – ReACT <br /> | ||
| - | **[[5xyj]], [[5xz5]] – yACT <br /> | ||
*Acetoacetyl-CoA thiolase complex | *Acetoacetyl-CoA thiolase complex | ||
| + | **[[1wl4]] – hACT cytosolic + CoA <br /> | ||
**[[1dm3]], [[1dlv]] – ZrACT + CoA<br /> | **[[1dm3]], [[1dlv]] – ZrACT + CoA<br /> | ||
**[[2wkt]], [[2wkv]], [[2wl4]], [[2wl5]], [[2vtz]], [[1m3z]] – ZrACT (mutant) + CoA<br /> | **[[2wkt]], [[2wkv]], [[2wl4]], [[2wl5]], [[2vtz]], [[1m3z]] – ZrACT (mutant) + CoA<br /> | ||
| Line 61: | Line 65: | ||
**[[2vu0]], [[1nl7]] - ZrACT + CoA<br /> | **[[2vu0]], [[1nl7]] - ZrACT + CoA<br /> | ||
**[[2vu1]], [[2vu2]], [[1ou6]] - ZrACT + pantheteine-11-pivalate<br /> | **[[2vu1]], [[2vu2]], [[1ou6]] - ZrACT + pantheteine-11-pivalate<br /> | ||
| - | **[[1wl4]] – hACT cytosolic + CoA <br /> | ||
**[[4xl4]] – CaACT + CoA <br /> | **[[4xl4]] – CaACT + CoA <br /> | ||
**[[6et9]] – MtACT + PFAM DUF35 + hydroxymethylglutaryl-CoA synthase - ''Methanothermococcus thermolithotrophicus''<br /> | **[[6et9]] – MtACT + PFAM DUF35 + hydroxymethylglutaryl-CoA synthase - ''Methanothermococcus thermolithotrophicus''<br /> | ||
**[[6esq]] – MtACT + CoA + PFAM DUF35 + hydroxymethylglutaryl-CoA synthase <br /> | **[[6esq]] – MtACT + CoA + PFAM DUF35 + hydroxymethylglutaryl-CoA synthase <br /> | ||
**[[6are]] – ACT + CoA + acetate - ''Aspergillus fumigatus'' <br /> | **[[6are]] – ACT + CoA + acetate - ''Aspergillus fumigatus'' <br /> | ||
| + | |||
| + | *SCP2-thiolase | ||
| + | |||
| + | **]]6hrv]] – zfSCP2 - zebrafish <br /> | ||
| + | **]]6hsj]] – zfSCP2 + CoA<br /> | ||
| + | **]]6hsp]] – zfSCP2 + CoA + octanoyl-CoA<br /> | ||
}} | }} | ||
== References == | == References == | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Revision as of 11:01, 20 February 2020
| |||||||||||
3D Structures of Thiolase
Updated on 20-February-2020
References
- ↑ Sundaramoorthy R, Micossi E, Alphey MS, Germain V, Bryce JH, Smith SM, Leonard GA, Hunter WN. The crystal structure of a plant 3-ketoacyl-CoA thiolase reveals the potential for redox control of peroxisomal fatty acid beta-oxidation. J Mol Biol. 2006 Jun 2;359(2):347-57. Epub 2006 Mar 29. PMID:16630629 doi:http://dx.doi.org/10.1016/j.jmb.2006.03.032
- ↑ Soto G, Stritzler M, Lisi C, Alleva K, Pagano ME, Ardila F, Mozzicafreddo M, Cuccioloni M, Angeletti M, Ayub ND. Acetoacetyl-CoA thiolase regulates the mevalonate pathway during abiotic stress adaptation. J Exp Bot. 2011 Nov;62(15):5699-711. doi: 10.1093/jxb/err287. Epub 2011 Sep 9. PMID:21908473 doi:http://dx.doi.org/10.1093/jxb/err287
- ↑ Kiema TR, Harijan RK, Strozyk M, Fukao T, Alexson SE, Wierenga RK. The crystal structure of human mitochondrial 3-ketoacyl-CoA thiolase (T1): insight into the reaction mechanism of its thiolase and thioesterase activities. Acta Crystallogr D Biol Crystallogr. 2014 Dec 1;70(Pt 12):3212-25. doi:, 10.1107/S1399004714023827. Epub 2014 Nov 22. PMID:25478839 doi:http://dx.doi.org/10.1107/S1399004714023827