We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1611
From Proteopedia
(Difference between revisions)
| Line 18: | Line 18: | ||
[https://www.nature.com/articles/s41594-018-0049-1 Nature_Structural_&_Molecular_Biology_Vol_25] | [https://www.nature.com/articles/s41594-018-0049-1 Nature_Structural_&_Molecular_Biology_Vol_25] | ||
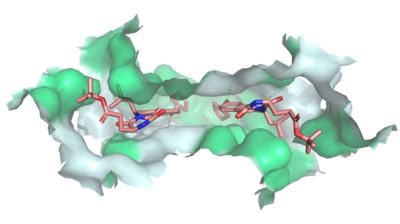
| - | Multidrug Transporter ABCG2 is a dimer that consists of two cavities seperated by a leucine plug. Cavity 1 is a binding pocket open to the cytoplasm and the inner leaflet of the plasma membrane. Its shape is suitable to bind flat, hydrophobic and polycyclic substrates. Many of its amino acids residues form hydrophobic interactions with the bound substrate, as shown in green in '''Figure 1'''. Cavity 2 is located above the leucine plug. It is empty until the substrate and ATP are bound and in place. Its inter- and intra-disulfides promote the release of the substrate from the cavity into the extracellular space. | + | Multidrug Transporter ABCG2 is a <scene name='83/832937/Dimer/1'>dimer</scene> that consists of two cavities seperated by a leucine plug. Cavity 1 is a binding pocket open to the cytoplasm and the inner leaflet of the plasma membrane. Its shape is suitable to bind flat, hydrophobic and polycyclic substrates. Many of its amino acids residues form hydrophobic interactions with the bound substrate, as shown in green in '''Figure 1'''. Cavity 2 is located above the leucine plug. It is empty until the substrate and ATP are bound and in place. Its inter- and intra-disulfides promote the release of the substrate from the cavity into the extracellular space. |
</StructureSection> | </StructureSection> | ||
Revision as of 03:22, 24 March 2020
| This Sandbox is Reserved from Jan 13 through September 1, 2020 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1598 through Sandbox Reserved 1627. |
To get started:
More help: Help:Editing |
Multidrug Transporter ABCG2 in Homo Sapiens
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
- ↑ Jackson SM, Manolaridis I, Kowal J, Zechner M, Taylor NMI, Bause M, Bauer S, Bartholomaeus R, Bernhardt G, Koenig B, Buschauer A, Stahlberg H, Altmann KH, Locher KP. Structural basis of small-molecule inhibition of human multidrug transporter ABCG2. Nat Struct Mol Biol. 2018 Apr;25(4):333-340. doi: 10.1038/s41594-018-0049-1. Epub, 2018 Apr 2. PMID:29610494 doi:http://dx.doi.org/10.1038/s41594-018-0049-1
Student Contributors
- Shelby Skaggs