This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1619
From Proteopedia
(New page: {{Sandbox_Reserved_CH462_Biochemistry_II}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> ==Your Heading Here (maybe something like 'Structure')== <StructureSection load='1stp' size='340' side...) |
|||
| Line 1: | Line 1: | ||
| - | + | ='''Gamma Secretase'''= | |
| - | = | + | <scene name='83/837256/Practice_scene_1/2'>Text To Be Displayed</scene>=''Human'' Gamma Secretase= |
| - | <StructureSection load=' | + | <StructureSection load='5A63' size='350' frame='true' side='right' caption='Gamma Secretase 5A63' scene='83/837256/Practice_scene_1/1'> |
| - | + | =Gamma Secretase= | |
| - | + | ||
| + | ==Introduction== | ||
| + | |||
| + | ===Background=== | ||
| + | Gamma Secretase is a transmembrane aspartate protease. It catalyzes peptide bond hydrolysis of type I integral membrane proteins such as Notch, APP, and various other substrates. It recognizes and catalyzes the reaction with its substrate using 3 residue segments. These substrates generate amyloid-β (Aβ). This product is important for various neural processes, and it is well known for its Implications with Alzheimer’s disease (AD). This has made gamma secretase a popular drug target, specifically using gamma secretase (GS) inhibitors. However, due to the nature of gamma secretase having various neural functions, there are dangerous side effects when it is inhibited. | ||
| + | |||
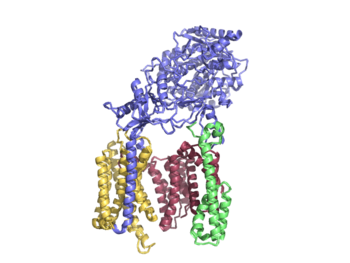
| + | ===Overall Structure=== | ||
| + | Γ-secretase is composed of 20 transmembrane components (TMs) and has 4 subunits: Nicastran, Anterior Pharynx-defective 1, Presenilin, and Presenilin Enhancer 2. These subunits are stabilized by hydrophobic interactions and 4 phosphatidylcholines.These phosphatidylcholines have interfaces between: PS1 and PEN-2, APH-1 and PS1, APH-1 and NCT. | ||
| + | Nicastrin (NCT) has a large extracellular domain and 1 TM. It is important to substrate recognition and binding. | ||
| + | Presenilin (PS1) serves as the active site of the protease and contains 9 TMs, each varying in length. The site of autocatalytic cleavage is located between TM6 and TM7 in PS1 and major conformational changes take place in this subunit upon substrate binding. | ||
| + | Anterior pharynx-defective 1 (APH-1) serves as a scaffold for anchoring and supporting the flexible conformational changes of PS1 | ||
| + | Activation of the active site is dependent on the binding of Presenilin enhancer 2 (PEN-2). PEN-2 is also important in maturation of the enzyme. | ||
| + | |||
| + | [[Image:Ribbon structure of whole.png|350 px|right|thumb|Figure 1]] | ||
| + | [http://http://proteopedia.org/wiki/index.php/5a63 5A63 Article] | ||
| + | ===practice=== | ||
| + | |||
| + | <scene name='83/837256/Practice_scene_1/2'>practice 2</scene> | ||
| - | == Function == | ||
| - | == Disease == | ||
| - | == Relevance == | ||
== Structural highlights == | == Structural highlights == | ||
| + | ===Substrate Structure=== | ||
| + | Though gamma secretase has multiple substrates, the substrate of main concern is called Amyloid Precursor Protein (APP). APP is composed of an N-terminal loop, a transmembrane (TM) helix, and a C-terminal β-strand. The uses lateral diffusion as a mechanism of entry into the enzyme, and once in place, the TM helix is anchored by hydrogen bonds. In order to differentiate substrates, the β-strand is often the main point of identification for the enzyme. After this, the helix undergoes unwinding and the process of catalysis can begin. | ||
| + | |||
| + | ===Lid Complex=== | ||
| + | Lid is the first point of entry and recognition for the substrate. | ||
| + | |||
| + | ===Active Site=== | ||
| + | The active site is located between TM6 and TM7 of the PS1 subunit, which is mainly hydrophilic and disordered. Each of these transmembrane helices has an aspartate residue, Asp257 and Asp385, which are located approximately 10.6 A˚ apart when inactive. | ||
| + | |||
| + | ==Relevance== | ||
| + | APP build up leads to Amyloid plaques in brain | ||
| + | Inhibition of γ-secretase could be potential AD treatment | ||
| + | Location of majority of mutations | ||
| + | Over 200 mutations that cause AD | ||
| + | AD mutations heavily target interface between PS1 and APP | ||
| + | Important to look at differences between substrates | ||
| + | |||
| + | |||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| + | 1.Bai X, Yan C, Guanghui Yang, et al. 2015. An atomic structure of human γ-secretase. Nature. 525:212-217. | ||
| + | 2.Carroll CM, and Li YM. 2016. Physiological and pathological roles of the γ-secretase complex. Brain research bulletin. 126:199-206. | ||
| + | 3.Yang G, Zhou R, Shi Y. 2017. Cryo-EM structures of human γ-secretase. Current Opinion in Structural Biology. 46:55–64. | ||
| + | 4.Yang G, Zhou R, Zhou Q, et al. 2019. Structural basis of Notch recognition by human γ-secretase. Nature. 565: 192-197. | ||
| + | 5.Zhou R, Yang G, Guo X, et al. 2019. Recognition of the amyloid precursor protein by human γ-secretase. Science. 363:1-8. | ||
<references/> | <references/> | ||
| + | |||
| + | ==Student Contributors== | ||
| + | Layla Wisser | ||
| + | Daniel Mulawa | ||
Revision as of 20:51, 24 March 2020
Gamma Secretase
=Human Gamma Secretase=
| |||||||||||
References
1.Bai X, Yan C, Guanghui Yang, et al. 2015. An atomic structure of human γ-secretase. Nature. 525:212-217. 2.Carroll CM, and Li YM. 2016. Physiological and pathological roles of the γ-secretase complex. Brain research bulletin. 126:199-206. 3.Yang G, Zhou R, Shi Y. 2017. Cryo-EM structures of human γ-secretase. Current Opinion in Structural Biology. 46:55–64. 4.Yang G, Zhou R, Zhou Q, et al. 2019. Structural basis of Notch recognition by human γ-secretase. Nature. 565: 192-197. 5.Zhou R, Yang G, Guo X, et al. 2019. Recognition of the amyloid precursor protein by human γ-secretase. Science. 363:1-8.
Student Contributors
Layla Wisser Daniel Mulawa