Introduction
The insulin receptor is a transmembrane protein receptor that is activated by the binding of insulin and is a vital proponent of cellular function. The insulin receptor belongs to the large class of tyrosine kinase receptors (RTKs). RTKs have a high affinity at the cell surface to bind to a particular ligand and are made up of three distinct parts: an extracellular domain with binding sites, a transmembrane region, and an intracellular domain with the tyrosine kinases that lead to downstream signaling upon activation of each other. The unique ability of the insulin receptor in particular to change conformation allows it to play a key role in a variety of cellular pathways including glucose homeostasis, regulation of lipid, protein, and carbohydrate metabolism, gene expression, and even modulation of brain neurotransmitter levels. This page focuses specifically on the insulin receptor's role in glucose homeostasis.
Structural Overview
The insulin receptor is a dimer of heterodimers made of two alpha subunits and two beta subunits [1].The are on the extracellular side of the membrane and are critical for binding insulin. The have the potential to interact with insulin ligands on the extracellular side of the membrane. There can be up to four binding sites, but it is generally more common for only one or two insulin molecules to bind to the receptor due to the occurrence of negative affinity at the binding site. The are transmembrane subunits that contain a tyrosine kinase region [1]. When the entire receptor experiences a conformation change from the V shape to the T shape upon activation or binding of an insulin molecule, the Beta chains are brought in close proximity to each other. When the two subunits are brought near to each other in the activated T form, the Tyrosine Kinase regions are able to autophosphorylate their counterparts at particular Tyrosine locations. It is important to note in the overall discussion of the insulin receptor structure that it has only been imaged in pieces, and not as a whole at this point in time. There are proposed structures of the entire molecule based off of the known function of the autophosphorylation, but the structure discussed throughout this page only contains part of the Beta subunits.
Function
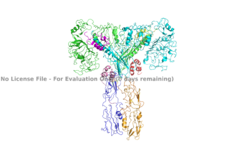
Figure 1. An image of the active insulin receptor showing both the alpha and beta subunits and insulin bound to the four binding sites. The dimers of the alpha subunit are colored in light blue and green. The intracellular beta sites are colored in blue and orange. The insulin binding sites with insulin bound are colored in magenta, light pink, yellow, and red.
PDB: 6SOF.
The insulin receptor's structure is critical to it's function. In regards to glucose homeostasis, the receptor begins the signaling pathway that will eventually move glucose transporters to the cell surface which will allow glucose to passively defuse into the cell. The glucose receptor is inactive in the absence of insulin. When insulin does bind to the receptor, it undergoes a conformation change from the inactive inverted V state to the active T state (Figure 1). Once activated, the intracellular Beta subunits move together close enough to autophosphorylate, and downstream signaling begins by the phosphorylation of the Insulin Receptor Substrate (IRS), ultimately resulting in glucose intake.
Studies have found that optimal insulin receptor activation requires the binding of multiple insulin ligands to two insulin binding sites. In (Figure 3) these two binding sites are colored in magenta and red. Binding of at least one insulin is required for the activation of the insulin receptor and the change in conformation to the active T state. [2].
Structure
The insulin receptor is a receptor tyrosine kinase that exists in two stable conformations, an inactive and active state. The entire insulin receptor is a dimer of heterodimers with two extracellular alpha subunits, and two transmembrane/intracellular beta subunits linked and stabilized by multiple disulfide bonds. The Cryo-EM structure of the extracellular domain of the Insulin Receptor without the presence of insulin bound at its alpha subunit site was established first, and is also known as the apo-form. The shape that it displayed appeared as an upside down V. Then, a subsequent Cryo-EM was established with insulin bound to the alpha subunit binding site, displaying a T shape conformation (UCHIKAWA).
Alpha Subunit
The alpha subunit is the extracellular domain of the insulin receptor and is the site of insulin binding. The subunit is unqiue in the way it binds insulin, and has four potential sites for binding. Each site performs binding via a "cross link", which is due to the fact that the receptor is a dimer of heterodimers and contains four protomers of similar structure. Each time an insulin ligand binds, the it comes in contact with the L1 domain of one protomer, and the alpha-CT chain and FnIII-1 loop of another protomer. This cross binding can occur in two sites (1 and 1'). There is potential for binding at sites 2 and 2', but it is less likely due to negative cooperation and the location of the sites on the back of the beta sheet of the FnIII-1 domain on each protomer. The alpha subunit is comprised of two Leucine rich domains (L1 & L2), a Cysteine rich domain (CR), and a C-terminal alpha helix. The actual site of insulin binding occurs at the and is stabilized by the L1 and L2 domains . The T shape conformation was also well observed in the alpha subunit. It is horizontally composed of L1, CR (including the alpha-CT chain), and L2 domains and vertically composed of the FnIII-1, 2, and 3 domains.
Beta Subunit
The beta subunit is the membrane spanning and intracellular portion of the insulin receptor. This domain is composed of three Fibronectin domains (FN III-1,-2,-3) and the tyrosine kinase domain. The tyrosine kinase domain is the site of autophosphorylation upon activation of the receptor.
Conformation Change
The inactive form of the insulin receptor predominates in low-levels of circulating insulin, whereas the active conformation is seen when insulin binds to any of the 4 receptor sites. The inactive conformation resembles an , and the active conformation resembles . The image of the inverted V conformation shows only a protomer of the inactive alpha subunit because the entire inactive alpha subunit dimer has been unable to be photographed because the transition state has yet to be determined in full. In the V-shape, the FnIII-3 domains are separated by about 120A. At this distance, they cannot work together to autophosphorylate. Upon the binding of insulin to any of the four binding sites, the conformation change will begin and bring the FnIII-3 domains within 40A of each other, which is the T-state conformation. Cryo-EM results have displayed clear representations of FnIII-2 and FnIII-3 domains, but lack in high density results for the transmembrane domain and cannot truly model anything past the two fibronectin domains due to the lack of side chain density. Due to the fact that FnIII-3 is connected to the transmembrane domain and intracellular kinase domains through a short linker, it is suggested that the insulin receptor does extend its T-shape conformation through the cell membrane and into the cell. Therefore, it is expected that the intracellular kinase domains will be in close proximity when this conformation change occurs extracellularly, ultimately allowing for autophosphorylation. [3]. (ALOT OF THIS WAS UCHIKAWA)
Relevance
As mentioned in the Introduction, the insulin receptor is relevant to numerous biological functions of the body. In order to address the receptors role in glucose homeostasis, it is important to discuss its relevance in disease. In a healthy, normal-functioning human, each cell's insulin receptors have the ability to bind to an abundance of insulin that can get released by the pancreas in response to a rise of blood glucose levels. Without properly functioning insulin receptors that can respond to increases in insulin, and therefore glucose, medical intervention is necessary for survival.
Disease
One of the most common diseases involving the insulin receptor in regards to glucose uptake and homeostasis is diabetes mellitus. There are two types of diabetes- which are referred to as type 1 and type 2 diabetes. Type 1 diabetes is classified as "insulin dependent" and is characterized by an inability for the body to produce insulin. This is most often the result of damage or insufficiency in the Islets of Langerhans in the pancreas. Type 2 diabetes is classified as "insulin independent" and is the result of the body producing insufficient amounts of insulin, or not responding to the insulin. This often occurs because of high blood-glucose levels. Both types of diabetes are often treated with insulin injections, and diet and lifestyle changes. [4] [5].
Treatment of Diabetes with Insulin
At the Cellular Level
In the human body, the conformation change from the inactive to active state upon insulin binding has a time constant of six minutes. Once insulin binds and the beta subunits are brought within close proximity, autophosphorylation of the beta subunits begins. Phosphorylation at these sites reaches a maximal level in about one minute, and lasts for approximately six to ten minutes. One insulin receptor substrate has a half-life of 3.5 minutes where it is able to be phosphorylated by the tyrosine kinases of the beta subunit and then act as a central hub to activate further downstream signaling pathways that eventually bring glucose receptors to the surface of the cell to allow for diffusion of glucose into the cell. Once insulin binds to the alpha subunit, the receptor remains active for approximately ten minutes before the insulin is degraded and the receptor returns to its inactive conformation. This time frame puts a perspective on how long it takes for the human body to store excess glucose in their blood stream from a recent meal as glycogen for later use as fuel.
An example for how to call up a reference in the future. You originally used this reference at the beginning of the Structural Overview. Now I have just taken the name in brackets and it will use the same number as the first time that this reference was used.
[1]

