We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Harrison L. Smith/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
==Structural Overview== | ==Structural Overview== | ||
| - | The insulin receptor is a [http://en.wikipedia.org/wiki/Dimer_(chemistry) dimer] of heterodimers made of two <scene name='83/832953/Alpha_subunits/1'>Alpha subunits</scene> and | + | The insulin receptor is a [http://en.wikipedia.org/wiki/Dimer_(chemistry) dimer] of heterodimers made of two <scene name='83/832953/Alpha_subunits/1'>Alpha subunits</scene> and <scene name='83/832953/Beta_subunits/1'>Beta subunits</scene> <ref name="Tatulian">PMID:26322622</ref>. There are four potential <scene name='83/832953/Binding_sites/1'>binding sites</scene> that can interact with insulin ligands on the extracellular side of the membrane. The structure of the insulin receptor can exist in two stable conformations, where it will be inactive and appear as an inverted V or active and appear as a right side up T shape. |
| - | + | ||
====Alpha Subunits==== | ====Alpha Subunits==== | ||
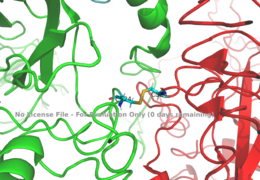
| - | The alpha subunits make up the extracellular domain ([http://en.wikipedia.org/wiki/Ectodomain ectodomain]) of the insulin receptor and are the sites of insulin binding. The alpha subunit is comprised of two Leucine rich domains (L1 & L2), a Cysteine rich domain (CR), and a C-terminal alpha helix. The actual site of insulin binding occurs at the <scene name='83/832953/Alpha_c_helix/1'>α-CT chain</scene> and is stabilized by the L1 and L2 domains <scene name='83/832953/Inactive_insulin_receptor/2'> Not sure what these are sos</scene>.There are two types of binding sites in the alpha subunits, Sites 1 and 1' and then Sites 2 and 2'. These two types have unique differences from each other, which makes the affinity for the first two sites, 1 and 1', much higher than that of sites 2 and 2'. The sites are in pairs because the receptor is a dimer of heterodimers and contains four protomers of similar structure. Each time an insulin ligand binds to sites 1 and 1', it comes in contact with the L1 domain of one protomer and the alpha-CT chain and FnIII-1 loop of another protomer, which is also known as "cross linking". There is potential for binding at sites 2 and 2', but it is less likely due to the location of the sites on the back of the beta sheet of the FnIII-1 domain on each protomer and the fact that there is a much greater surface area for binding at sites 1 and 1'. <ref name="Uchikawa"> DOI 10.7554/eLife.48630 </ref>. | + | [[Image:Disulfide bridge between alphas.png|thumb|right|260px|Figure 1: Disulfide bridge between the two alpha subunits. [http://www.rcsb.org/structure/6sof PDB 6SOF]]] |
| - | ===Beta | + | The alpha subunits make up the extracellular domain ([http://en.wikipedia.org/wiki/Ectodomain ectodomain]) of the insulin receptor and are the sites of insulin binding. The alpha subunit is comprised of two Leucine rich domains (L1 & L2), a Cysteine rich domain (CR), and a C-terminal alpha helix. The two subunits are held together by a [http://en.wikipedia.org/wiki/Disulfide disulfide bond] at residue C524 of one alpha subunit, and C524 of the other subunit (Figure1). The actual site of insulin binding occurs at the <scene name='83/832953/Alpha_c_helix/1'>α-CT chain</scene> and is stabilized by the L1 and L2 domains <scene name='83/832953/Inactive_insulin_receptor/2'> Not sure what these are sos</scene>.There are two types of binding sites in the alpha subunits, Sites 1 and 1' and then Sites 2 and 2'. These two types have unique differences from each other, which makes the affinity for the first two sites, 1 and 1', much higher than that of sites 2 and 2'. The sites are in pairs because the receptor is a dimer of heterodimers and contains four protomers of similar structure. Each time an insulin ligand binds to sites 1 and 1', it comes in contact with the L1 domain of one protomer and the alpha-CT chain and FnIII-1 loop of another protomer, which is also known as "cross linking". There is potential for binding at sites 2 and 2', but it is less likely due to the location of the sites on the back of the beta sheet of the FnIII-1 domain on each protomer and the fact that there is a much greater surface area for binding at sites 1 and 1'. <ref name="Uchikawa"> DOI 10.7554/eLife.48630 </ref>. |
| - | The beta subunit is part of the extracellular membrane as well, but also spans the membrane region and inserts the intracellular portion of the insulin receptor. This domain is composed of part of [ | + | ===Beta Subunits=== |
| + | The beta subunit is part of the extracellular membrane as well, but also spans the membrane region and inserts the intracellular portion of the insulin receptor. This domain is composed of part of [http://en.wikipedia.org/wiki/Fibronectin fibronectin] domain III-2 and all of Fibronectin domain III-3. It is proposed that the beta subunit's FnIII-3 domain has links through the transmembrane region into the intracellular part of the membrane. Due to the fact that it is known that there is a conformation change in the FnIII-3 extracellular part, it is expected that the conformation change follows all the way through to the end of the receptor in order to accomplish the complete T-shape which will ultimately induce the autophosphorylation of the tyrosine kinase domain. | ||
===Cryo-EM Structural Imaging=== | ===Cryo-EM Structural Imaging=== | ||
| - | [ | + | [http://en.wikipedia.org/wiki/Transmission_electron_cryomicroscopy Cryo-EM] results have displayed clear representations of FnIII-2 and FnIII-3 domains, but lack in high density results for the transmembrane domain and cannot truly model anything past the two fibronectin domains due to the lack of side chain density. Due to the fact that FnIII-3 is connected to the transmembrane domain and intracellular kinase domains through a short linker, it is suggested that the insulin receptor does extend its T-shape conformation through the cell membrane and into the cell. Therefore, it is expected that the intracellular kinase domains will be in close proximity when this conformation change occurs extracellularly, ultimately allowing for [http://en.wikipedia.org/wiki/Autophosphorylation autophosphorylation]. The Cryo-EM structure of the extracellular domain of the Insulin Receptor without the presence of insulin bound at its alpha subunit site was established first, and is also known as the apo-form. The shape that it displayed appeared as an upside down V. Then, a subsequent Cryo-EM was established with insulin bound to the alpha subunit binding site, displaying a T shape conformation of the same alpha protomer unit.<ref name="Uchikawa" /> It is important to note in the overall discussion of the insulin receptor structure that it has only been imaged in pieces, and not as a whole at this point in time. There are proposed structures of the entire molecule based off of the known function of the tyrosine autophosphorylation and downstream activation, but the structure discussed throughout this page only contains part of the Beta subunits through that of the FnIII-3 domain. To analyze work completed on the tyrosine kinase domain of the receptor, [https//www.rcsb.org/structure/1ir3 PDB 1IR3] can be referenced. |
===Insulin=== | ===Insulin=== | ||
| - | [[Image: | + | [[Image:Purple insulin.png|thumb|right|150px|Figure 2: Insulin molecule [http://www.rcsb.org/structure/3I40 PDB 3I40]]]Insulin is a [http://en.wikipedia.org/wiki/Peptide_hormone peptide hormone] produced and secreted from the [http://en.wikipedia.org/wiki/Pancreatic_islets islets of Langerhans] of the pancreas in response to high blood glucose levels. Insulin is commonly considered the anabolic hormone of the body, and is the only [http://en.wikipedia.org/wiki/Ligand ligand] that is capable of binding to and activating the insulin receptor. The structure of insulin is fairly simple, it is a monomer composed of two peptide chains whcih are linked together by a disulfide bridge. In regards to glucose homeostasis, insulin is needed to begin the process of bringing extracellular glucose into the cell to be converted into [http://en.wikipedia.org/wiki/Glycogen glycogen] for storage and later usage. |
| + | |||
== Function== | == Function== | ||
| - | + | The insulin receptor's structure is critical to it's function. In regards to glucose homeostasis, the receptor begins the signaling pathway that will eventually move glucose transporters to the cell surface which will allow glucose to passively defuse into the cell. The glucose receptor is inactive in the absence of insulin. When a large meal is ingested and there is a surplus of glucose circulating in the blood stream, the production of insulin is upregulated and will bind to many insulin receptors. Upon activation, it undergoes a structural [http://en.wikipedia.org/wiki/Conformational_change conformation change] from the inactive <scene name='83/832953/Simple_inactivated_receptor/1'>inverted V</scene> state to the active <scene name='83/832953/Ir_dimer_t_state/1'>T shape</scene> state. Once activated, the Beta subunits are enabled to move close enough together to autophosphorylate and initate downstream signaling by the phosphorylation of the [http://en.wikipedia.org/wiki/Insulin_receptor_substrate Insulin Receptor Substrate] (IRS), ultimately resulting in glucose intake. | |
| - | The insulin receptor's structure is critical to it's function. In regards to glucose homeostasis, the receptor begins the signaling pathway that will eventually move glucose transporters to the cell surface which will allow glucose to passively defuse into the cell. The glucose receptor is inactive in the absence of insulin. When a large meal is ingested and there is a surplus of glucose circulating in the blood stream, the production of insulin is upregulated and will bind to many insulin receptors. Upon activation, it undergoes a structural [ | + | |
===Conformation Change=== | ===Conformation Change=== | ||
The inactive form of the insulin receptor predominates in low-levels of circulating insulin, whereas the active conformation is seen when insulin binds to any of the 4 receptor sites. The inactive conformation resembles an inverted V, and the active conformation resembles a T shape. Images of the inverted V conformation shows only a protomer of the inactive alpha subunit because the entire inactive alpha subunit dimer has been unable to be photographed because the transition state has yet to be determined in full. In the V-shape, the FnIII-3 domains are separated by about 120A. At this distance, they cannot work together to autophosphorylate and initiate downstream signaling. Upon the binding of insulin to any of the four binding sites, the conformation change will begin and bring the FnIII-3 domains within 40A of each other, which is the T-state conformation. <ref> DOI 10.1038/s41467-018-06826-6</ref> <ref name="Uchikawa" /> The T shape conformation is well observed in the alpha subunit. It is horizontally composed of L1, CR (including the alpha-CT chain), and L2 domains and vertically composed of the FnIII-1, 2, and 3 domains. | The inactive form of the insulin receptor predominates in low-levels of circulating insulin, whereas the active conformation is seen when insulin binds to any of the 4 receptor sites. The inactive conformation resembles an inverted V, and the active conformation resembles a T shape. Images of the inverted V conformation shows only a protomer of the inactive alpha subunit because the entire inactive alpha subunit dimer has been unable to be photographed because the transition state has yet to be determined in full. In the V-shape, the FnIII-3 domains are separated by about 120A. At this distance, they cannot work together to autophosphorylate and initiate downstream signaling. Upon the binding of insulin to any of the four binding sites, the conformation change will begin and bring the FnIII-3 domains within 40A of each other, which is the T-state conformation. <ref> DOI 10.1038/s41467-018-06826-6</ref> <ref name="Uchikawa" /> The T shape conformation is well observed in the alpha subunit. It is horizontally composed of L1, CR (including the alpha-CT chain), and L2 domains and vertically composed of the FnIII-1, 2, and 3 domains. | ||
| + | |||
===Binding interactions=== | ===Binding interactions=== | ||
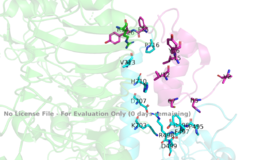
| - | + | [[Image:Binding site with AA labeled.png|thumb|right|270px|Figure 3: Subunit interactions between the insulin receptor CT-alpha helix (light blue) and insulin (magenta) in one of the binding sites [http://www.rcsb.org/structure/6sof PDB 6SOF]]] | |
| + | The insulin receptor itself is held together by numerous critical disulfide bonds and [http://en.wikipedia.org/wiki/Salt_bridge_(protein_and_supramolecular) salt bridges]. These bonds maintain a stablized link between the dimers of the receptor, and without them, the conformation change from inactive to active would not be able to occur. One unique interaction within the receptor is known as a tripartite interaction. It occurs between the alpha-CT chain and the FnIII-1 domain region during a conformation chain, and involves the following residues:<scene name='83/832953/Alpha_ct_and_fniii-1/2'> ASP496, ARG498, and ASP99 on the FnIII domain</scene> and the <scene name='83/832953/Alpha_ct_and_fniii-1/3'>LYS703, GLU706, and ASP707 on the alpha-CT domain</scene> . This duo then interacts with the leucine rich region, L1, that exists on the opposing protomer of the dimer. | ||
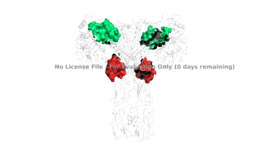
| + | [[Image:4 sites highlighted.png|thumb|right|260px|Figure 4: Sites 1, 1', 2, and 2'. [http://www.rcsb.org/structure/6sof PDB 6SOF]]] | ||
| + | It is generally more common for only one or two insulin molecules to bind to the receptor due to the occurrence of negative affinity at the binding site, as well as the location of the second two binding sites on the back side of the receptor with Beta sheets and the lack of surface area they have. For this reason, studies support that optimal insulin receptor activation requires the binding of ligands to two insulin binding sites. Binding of at least one insulin is required for the activation of the insulin receptor and the change in conformation to the active T state. <ref> DOI 10.7554/eLife.48630 </ref>. | ||
| - | + | Ligand binding interactions are different at sites 1 and 1' and 2 and 2'. Sites 1 and 1' are highlighted in green and sites 2 and 2' are highlighted in red (Figure 4). The interactions at all 4 binding sites are highly hydrophobic, but there are differences between the sites. _______ have two cystines in a disulfide bond linkage along with HIS B5 all from insulin interacting with proline, phenylalanine, and arginine residues from the FnIII-1 domain. <scene name='83/832953/Sites_2_and_2_prime_location/1'>Sites 2 and 2'</scene>experience an interaction with insulin that involves more residues than at the first two sites. The FnIII-1 region has both basic residues-ARG479, LYS484, ARG488, ARG554- and hydrophobic residues- LEU486, LEU552, and PRO537- interacting with insulins resides and numerous locations along its surface. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | Ligand binding interactions are different at sites 1 and 1' and 2 and 2'. | + | |
| - | 1 and 1' | + | |
| - | 2 and 2' | + | |
== Relevance == | == Relevance == | ||
As mentioned in the Introduction, the insulin receptor is relevant to numerous biological functions of the body. In order to address the receptors role in glucose homeostasis, it is important to discuss its relevance in disease. In a healthy, normal-functioning human, each cell's insulin receptors have the ability to bind to an abundance of insulin that can get released by the pancreas in response to a rise of blood glucose levels. Without properly functioning insulin receptors that can respond to increases in insulin, and therefore glucose, medical intervention is necessary for survival. | As mentioned in the Introduction, the insulin receptor is relevant to numerous biological functions of the body. In order to address the receptors role in glucose homeostasis, it is important to discuss its relevance in disease. In a healthy, normal-functioning human, each cell's insulin receptors have the ability to bind to an abundance of insulin that can get released by the pancreas in response to a rise of blood glucose levels. Without properly functioning insulin receptors that can respond to increases in insulin, and therefore glucose, medical intervention is necessary for survival. | ||
=== Disease === | === Disease === | ||
| - | One of the most common diseases involving the insulin receptor in regards to glucose uptake and homeostasis is [ | + | One of the most common diseases involving the insulin receptor in regards to glucose uptake and homeostasis is [http://en.wikipedia.org/wiki/Diabetes diabetes mellitus]. There are two types of diabetes- which are referred to as type 1 and type 2 diabetes. Type 1 diabetes is classified as "insulin dependent" and is characterized by an inability for the body to produce insulin. This is most often the result of damage or insufficiency in the Islets of Langerhans in the pancreas. Type 2 diabetes is classified as "insulin independent" and is the result of the body producing insufficient amounts of insulin, or not responding to the insulin. This often occurs because of high blood-glucose levels. Both types of diabetes are often treated with insulin injections, and diet and lifestyle changes. <ref name="Wilcox"> PMID:16278749</ref> <ref name= "Riddle"> PMID: 6351440</ref>. |
===At the Cellular Level=== | ===At the Cellular Level=== | ||
| Line 50: | Line 48: | ||
| - | + | ||
| - | + | ||
Revision as of 01:22, 7 April 2020
Homo sapiens Insulin Receptor
| |||||||||||
References
- ↑ 1.0 1.1 Tatulian SA. Structural Dynamics of Insulin Receptor and Transmembrane Signaling. Biochemistry. 2015 Sep 15;54(36):5523-32. doi: 10.1021/acs.biochem.5b00805. Epub , 2015 Sep 3. PMID:26322622 doi:http://dx.doi.org/10.1021/acs.biochem.5b00805
- ↑ 2.0 2.1 2.2 Uchikawa E, Choi E, Shang G, Yu H, Bai XC. Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex. Elife. 2019 Aug 22;8. pii: 48630. doi: 10.7554/eLife.48630. PMID:31436533 doi:http://dx.doi.org/10.7554/eLife.48630
- ↑ Weis F, Menting JG, Margetts MB, Chan SJ, Xu Y, Tennagels N, Wohlfart P, Langer T, Muller CW, Dreyer MK, Lawrence MC. The signalling conformation of the insulin receptor ectodomain. Nat Commun. 2018 Oct 24;9(1):4420. doi: 10.1038/s41467-018-06826-6. PMID:30356040 doi:http://dx.doi.org/10.1038/s41467-018-06826-6
- ↑ Uchikawa E, Choi E, Shang G, Yu H, Bai XC. Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex. Elife. 2019 Aug 22;8. pii: 48630. doi: 10.7554/eLife.48630. PMID:31436533 doi:http://dx.doi.org/10.7554/eLife.48630
- ↑ Wilcox G. Insulin and insulin resistance. Clin Biochem Rev. 2005 May;26(2):19-39. PMID:16278749
- ↑ Riddle MC. Treatment of diabetes with insulin. From art to science. West J Med. 1983 Jun;138(6):838-46. PMID:6351440
Student Contributors
- Harrison Smith
- Alyssa Ritter