We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1607
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
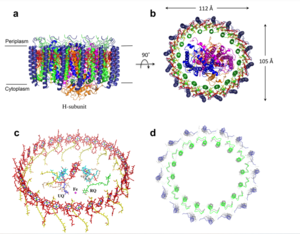
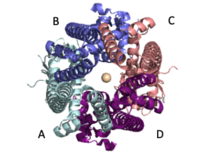
The selectivity pore is an integral part of the protein. This pore contains a group of glutamate with oxygen facing inward forming a carboxylate ring through which calcium enters. This negative carboxylate ring does a good job of pulling the positive calcium into the selectivity pore at the top of the protein. | The selectivity pore is an integral part of the protein. This pore contains a group of glutamate with oxygen facing inward forming a carboxylate ring through which calcium enters. This negative carboxylate ring does a good job of pulling the positive calcium into the selectivity pore at the top of the protein. | ||
| - | [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy Cryogenic electron microscopy] (Cryo-EM) was instrumental in outlining the complete structure of this protein. | + | [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy Cryogenic electron microscopy] (Cryo-EM) was instrumental in outlining the complete structure of this protein. <ref>DOI 10.1038/s41580-018-0052-8</ref> |
== Structural highlights and mechanism == | == Structural highlights and mechanism == | ||
| Line 22: | Line 22: | ||
===Selectivity Filter=== | ===Selectivity Filter=== | ||
| - | The <scene name='83/832933/Selectivity_filter/3'>selectivity filter</scene> contains Glu358, Trp354, and Pro359 to allow calcium to pass through the uniporter. The carboxylate oxygen of the <scene name='83/832933/Glu_358/2'>Glu358</scene> side chains draw in the positive calcium ion. The <scene name='83/832933/Diameter/1'>diameter</scene> of the carboxyl ring is about 4Å, allowing only a dehydrated Ca2+ ion to bind. Trp38, which is directly next to the Glu residues, stabilizes the carbonyl side chains through <scene name='83/832933/H_bond_trp354_glu358/2'>hydrogen bonding</scene> and anion pi interactions. These Trp residues also form stacking interactions with Pro359, which orientate the Glu carboxyl side chains towards the middle of the pore to interact with Ca2+ ions. | + | The <scene name='83/832933/Selectivity_filter/3'>selectivity filter</scene> contains Glu358, Trp354, and Pro359 to allow calcium to pass through the uniporter. The carboxylate oxygen of the <scene name='83/832933/Glu_358/2'>Glu358</scene> side chains draw in the positive calcium ion. The <scene name='83/832933/Diameter/1'>diameter</scene> of the carboxyl ring is about 4Å, allowing only a dehydrated Ca2+ ion to bind. Trp38, which is directly next to the Glu residues, stabilizes the carbonyl side chains through <scene name='83/832933/H_bond_trp354_glu358/2'>hydrogen bonding</scene> and anion pi interactions. These Trp residues also form stacking interactions with Pro359, which orientate the Glu carboxyl side chains towards the middle of the pore to interact with Ca2+ ions. <ref name=”Yoo”>PMID:29954988</ref> |
[http://www.rcsb.org/structure/6DT0 Calcium Uniporter Structure] | [http://www.rcsb.org/structure/6DT0 Calcium Uniporter Structure] | ||
| - | |||
| - | <ref name=”Yoo”>PMID:29954988</ref> | ||
== Function == | == Function == | ||
Revision as of 04:20, 7 April 2020
Mitochondrial Calcium Uniporter
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Giorgi C, Marchi S, Pinton P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat Rev Mol Cell Biol. 2018 Nov;19(11):713-730. doi: 10.1038/s41580-018-0052-8. PMID:30143745 doi:http://dx.doi.org/10.1038/s41580-018-0052-8
- ↑ Yoo J, Wu M, Yin Y, Herzik MA Jr, Lander GC, Lee SY. Cryo-EM structure of a mitochondrial calcium uniporter. Science. 2018 Jun 28. pii: science.aar4056. doi: 10.1126/science.aar4056. PMID:29954988 doi:http://dx.doi.org/10.1126/science.aar4056