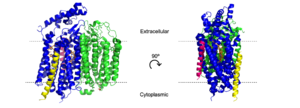We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1625
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
[[Image:Transmem1.png|300 px|right|thumb|'''Figure 2''. Cartoon model of cytochrome bd-oxidase in ''E. coli''. Dashed lines represent borders of cytoplasmic and extracellular regions.]] | [[Image:Transmem1.png|300 px|right|thumb|'''Figure 2''. Cartoon model of cytochrome bd-oxidase in ''E. coli''. Dashed lines represent borders of cytoplasmic and extracellular regions.]] | ||
| - | bd Oxidase is a type of quinol-dependent terminal oxidase found exclusively in prokaryotes.<ref name="Safarian">PMID: 27126043</ref> With a very high oxygen affinity, bd oxidases play a vital role in the oxidative phosphorylation pathway in both gram-positive and gram-negative bacteria. bd oxidases responsibility in the oxidative phosphorylation pathway allows the protein to also assist as a key survival factor in the bacterial stress response against antibacterial drugs. The <scene name='83/832931/Full/4'>cytochrome ''bd'' oxidase</scene> allows bacteria to be resistant to hypoxia, cyanide, nitric oxide, and H<sub>2</sub>O<sub>2</sub><ref name="Harikishore">PMID: 31939065</ref> | + | bd Oxidase is a type of quinol-dependent terminal oxidase found exclusively in prokaryotes.<ref name="Safarian">PMID: 27126043</ref> With a very high oxygen affinity, bd oxidases play a vital role in the oxidative phosphorylation pathway in both gram-positive and gram-negative bacteria. bd oxidases responsibility in the oxidative phosphorylation pathway allows the protein to also assist as a key survival factor in the bacterial stress response against antibacterial drugs. <ref name="Safarian">PMID: 31604309</ref> The <scene name='83/832931/Full/4'>cytochrome ''bd'' oxidase</scene> allows bacteria to be resistant to hypoxia, cyanide, nitric oxide, and H<sub>2</sub>O<sub>2</sub><ref name="Harikishore">PMID: 31939065</ref> |
Given this knowledge, bd oxidases have become an area of scientific research worth pursuing as they could serve as an ideal target for antimicrobial drug development. | Given this knowledge, bd oxidases have become an area of scientific research worth pursuing as they could serve as an ideal target for antimicrobial drug development. | ||
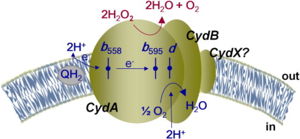
| - | [[Image:proton graadient.jpg|300 px|left|thumb|Figure 1: Overall schematic representation of cytochrome bd | + | [[Image:proton graadient.jpg|300 px|left|thumb|Figure 1: Overall schematic representation of cytochrome bd. <ref name= "Giuffre">PMID: 24486503</ref>; General display of the reduction of molecular oxygen into water using the quinol as a reducing substrate. The three hemes are located near the periplasmic space, meaning that the membrane potential is generated mainly from proton transfer from the cytoplasm towards the active site on the opposite site of the membrane. Heme''b558'' is involved in quinol oxidation and Heme''d'' serves as the site where O2 binds and becomes reduced to H2O.]] |
The overall mechanism of bd oxidases involves an exergonic reduction reaction of molecular oxygen into water (Figure 1). During this reaction, a proton gradient is generated in order to assist in the conservation of energy. Unlike other terminal oxidases, bd oxidases do not use a proton pump. Instead, bd oxidases use a form of vectorial chemistry that releases protons from the quinol oxidation into the positive, periplasmic side of the membrane. Protons that are required for the water formation are then consumed from the negative, cytoplasmic side of the membrane, thus creating the previously mentioned proton gradient. | The overall mechanism of bd oxidases involves an exergonic reduction reaction of molecular oxygen into water (Figure 1). During this reaction, a proton gradient is generated in order to assist in the conservation of energy. Unlike other terminal oxidases, bd oxidases do not use a proton pump. Instead, bd oxidases use a form of vectorial chemistry that releases protons from the quinol oxidation into the positive, periplasmic side of the membrane. Protons that are required for the water formation are then consumed from the negative, cytoplasmic side of the membrane, thus creating the previously mentioned proton gradient. | ||
Revision as of 04:51, 7 April 2020
| This Sandbox is Reserved from Jan 13 through September 1, 2020 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1598 through Sandbox Reserved 1627. |
To get started:
More help: Help:Editing |
Cytochrome bd-1 oxidase in Escherichia coli
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 Safarian S, Rajendran C, Muller H, Preu J, Langer JD, Ovchinnikov S, Hirose T, Kusumoto T, Sakamoto J, Michel H. Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases. Science. 2016 Apr 29;352(6285):583-6. doi: 10.1126/science.aaf2477. PMID:27126043 doi:http://dx.doi.org/10.1126/science.aaf2477
- ↑ 2.0 2.1 Harikishore A, Chong SSM, Ragunathan P, Bates RW, Gruber G. Targeting the menaquinol binding loop of mycobacterial cytochrome bd oxidase. Mol Divers. 2020 Jan 14. pii: 10.1007/s11030-020-10034-0. doi:, 10.1007/s11030-020-10034-0. PMID:31939065 doi:http://dx.doi.org/10.1007/s11030-020-10034-0
- ↑ Giuffre A, Borisov VB, Arese M, Sarti P, Forte E. Cytochrome bd oxidase and bacterial tolerance to oxidative and nitrosative stress. Biochim Biophys Acta. 2014 Jul;1837(7):1178-87. doi:, 10.1016/j.bbabio.2014.01.016. Epub 2014 Jan 31. PMID:24486503 doi:http://dx.doi.org/10.1016/j.bbabio.2014.01.016
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 Thesseling A, Rasmussen T, Burschel S, Wohlwend D, Kagi J, Muller R, Bottcher B, Friedrich T. Homologous bd oxidases share the same architecture but differ in mechanism. Nat Commun. 2019 Nov 13;10(1):5138. doi: 10.1038/s41467-019-13122-4. PMID:31723136 doi:http://dx.doi.org/10.1038/s41467-019-13122-4
- ↑ 5.0 5.1 5.2 5.3 5.4 Safarian S, Rajendran C, Muller H, Preu J, Langer JD, Ovchinnikov S, Hirose T, Kusumoto T, Sakamoto J, Michel H. Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases. Science. 2016 Apr 29;352(6285):583-6. doi: 10.1126/science.aaf2477. PMID:27126043 doi:http://dx.doi.org/10.1126/science.aaf2477
- ↑ 6.0 6.1 Hughes ER, Winter MG, Duerkop BA, Spiga L, Furtado de Carvalho T, Zhu W, Gillis CC, Buttner L, Smoot MP, Behrendt CL, Cherry S, Santos RL, Hooper LV, Winter SE. Microbial Respiration and Formate Oxidation as Metabolic Signatures of Inflammation-Associated Dysbiosis. Cell Host Microbe. 2017 Feb 8;21(2):208-219. doi: 10.1016/j.chom.2017.01.005. PMID:28182951 doi:http://dx.doi.org/10.1016/j.chom.2017.01.005
- ↑ 7.0 7.1 Shepherd M, Achard ME, Idris A, Totsika M, Phan MD, Peters KM, Sarkar S, Ribeiro CA, Holyoake LV, Ladakis D, Ulett GC, Sweet MJ, Poole RK, McEwan AG, Schembri MA. The cytochrome bd-I respiratory oxidase augments survival of multidrug-resistant Escherichia coli during infection. Sci Rep. 2016 Oct 21;6:35285. doi: 10.1038/srep35285. PMID:27767067 doi:http://dx.doi.org/10.1038/srep35285
- ↑ Arora K, Ochoa-Montano B, Tsang PS, Blundell TL, Dawes SS, Mizrahi V, Bayliss T, Mackenzie CJ, Cleghorn LA, Ray PC, Wyatt PG, Uh E, Lee J, Barry CE 3rd, Boshoff HI. Respiratory flexibility in response to inhibition of cytochrome C oxidase in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2014 Nov;58(11):6962-5. doi: 10.1128/AAC.03486-14., Epub 2014 Aug 25. PMID:25155596 doi:http://dx.doi.org/10.1128/AAC.03486-14
- ↑ Galvan AE, Chalon MC, Rios Colombo NS, Schurig-Briccio LA, Sosa-Padilla B, Gennis RB, Bellomio A. Microcin J25 inhibits ubiquinol oxidase activity of purified cytochrome bd-I from Escherichia coli. Biochimie. 2019 May;160:141-147. doi: 10.1016/j.biochi.2019.02.007. Epub 2019 Feb, 19. PMID:30790617 doi:http://dx.doi.org/10.1016/j.biochi.2019.02.007
- ↑ Lu P, Heineke MH, Koul A, Andries K, Cook GM, Lill H, van Spanning R, Bald D. The cytochrome bd-type quinol oxidase is important for survival of Mycobacterium smegmatis under peroxide and antibiotic-induced stress. Sci Rep. 2015 May 27;5:10333. doi: 10.1038/srep10333. PMID:26015371 doi:http://dx.doi.org/10.1038/srep10333
Student Contributors
- Grace Bassler
- Emily Neal
- Marisa Villarreal