We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1627
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
==Structural Overview== | ==Structural Overview== | ||
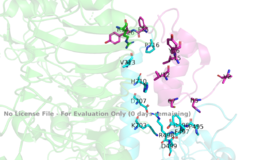
| - | The insulin receptor is a [http://en.wikipedia.org/wiki/Dimer_(chemistry) dimer] of heterodimers made of two <<scene name='83/832953/Alpha_subunits/4'>alpha subunits</scene> and two <scene name='83/832953/Beta_subunits/3'>beta subunits</scene> <ref name="Tatulian">PMID:26322622</ref>. | + | The insulin receptor is a [http://en.wikipedia.org/wiki/Dimer_(chemistry) dimer] of heterodimers made of two <<scene name='83/832953/Alpha_subunits/4'>alpha subunits</scene> and two <scene name='83/832953/Beta_subunits/3'>beta subunits</scene> <ref name="Tatulian">PMID:26322622</ref>. Within the extracellular ectodomain, there are four potential <scene name='83/832953/Binding_sites/2'>binding sites</scene> that can interact with insulin ligands on the extracellular side of the membrane. Binding of insulin initiates a large structural transition from an inactive, inverted V shape or an active T shape. |
====Alpha Subunits==== | ====Alpha Subunits==== | ||
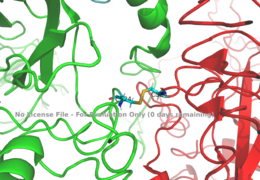
[[Image:Disulfide bridge between alphas.png|thumb|right|260px|Figure 1: Disulfide bridge (yellow) made of two cysteine residues (blue) that provides a linkage and stability to the two alpha subunits. [http://www.rcsb.org/structure/6sof PDB 6SOF]]] | [[Image:Disulfide bridge between alphas.png|thumb|right|260px|Figure 1: Disulfide bridge (yellow) made of two cysteine residues (blue) that provides a linkage and stability to the two alpha subunits. [http://www.rcsb.org/structure/6sof PDB 6SOF]]] | ||
| - | The alpha subunits make up the extracellular domain ([http://en.wikipedia.org/wiki/Ectodomain ectodomain]) of the insulin receptor and are the sites of insulin binding. The alpha subunit is comprised of two Leucine rich domains (L1 & L2), a Cysteine rich domain (CR), and a C-terminal alpha helix. The | + | The alpha subunits make up the extracellular domain ([http://en.wikipedia.org/wiki/Ectodomain ectodomain]) of the insulin receptor and are the sites of insulin binding. The alpha subunit is comprised of two Leucine rich domains (L1 & L2), a Cysteine rich domain (CR), and a C-terminal alpha helix. The alpha and beta subunits are held together by a [http://en.wikipedia.org/wiki/Disulfide disulfide bond] at residue C524 of one alpha subunit, and C524 of the other subunit (Figure1). The actual site of insulin binding occurs at the <scene name='83/832953/Alpha_c_helix/3'>α-CT chain</scene> of one of the sites and is stabilized by the L1 and L2 domains. Two types of binding sites in the alpha subunits, Sites 1 and 1' and then Sites 2 and 2'. These two types have unique differences from each other, which makes the affinity for the first two sites, 1 and 1', much higher than that of sites 2 and 2'. The sites are in pairs because the receptor is a dimer of heterodimers and contains four protomers of similar structure. Each time an insulin ligand binds to sites 1 and 1', it comes in contact with the L1 domain of one protomer and the alpha-CT chain and FnIII-1 loop of another protomer, which is also known as "cross linking". There is potential for binding at sites 2 and 2', but it is less likely due to the location of the sites on the back of the beta sheet of the FnIII-1 domain on each protomer and the fact that there is a much greater surface area for binding at sites 1 and 1'. <ref name="Uchikawa"> DOI 10.7554/eLife.48630 </ref>. |
===Beta Subunits=== | ===Beta Subunits=== | ||
The beta subunit is part of the extracellular membrane as well, but also spans the membrane region and inserts the intracellular portion of the insulin receptor. This domain is composed of part of [http://en.wikipedia.org/wiki/Fibronectin fibronectin] domain III-2 and all of Fibronectin domain III-3. It is proposed that the beta subunit's FnIII-3 domain has links through the transmembrane region into the intracellular part of the membrane. Due to the fact that it is known that there is a conformation change in the FnIII-3 extracellular part, it is expected that the conformation change follows all the way through to the end of the receptor in order to accomplish the complete T-shape which will ultimately induce the autophosphorylation of the tyrosine kinase domain. | The beta subunit is part of the extracellular membrane as well, but also spans the membrane region and inserts the intracellular portion of the insulin receptor. This domain is composed of part of [http://en.wikipedia.org/wiki/Fibronectin fibronectin] domain III-2 and all of Fibronectin domain III-3. It is proposed that the beta subunit's FnIII-3 domain has links through the transmembrane region into the intracellular part of the membrane. Due to the fact that it is known that there is a conformation change in the FnIII-3 extracellular part, it is expected that the conformation change follows all the way through to the end of the receptor in order to accomplish the complete T-shape which will ultimately induce the autophosphorylation of the tyrosine kinase domain. | ||
Revision as of 21:13, 14 April 2020
Homo sapiens Insulin Receptor
| |||||||||||
References
- ↑ 1.0 1.1 De Meyts P. The Insulin Receptor and Its Signal Transduction Network PMID:27512793
- ↑ 2.0 2.1 Tatulian SA. Structural Dynamics of Insulin Receptor and Transmembrane Signaling. Biochemistry. 2015 Sep 15;54(36):5523-32. doi: 10.1021/acs.biochem.5b00805. Epub , 2015 Sep 3. PMID:26322622 doi:http://dx.doi.org/10.1021/acs.biochem.5b00805
- ↑ 3.0 3.1 3.2 Uchikawa E, Choi E, Shang G, Yu H, Bai XC. Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex. Elife. 2019 Aug 22;8. pii: 48630. doi: 10.7554/eLife.48630. PMID:31436533 doi:http://dx.doi.org/10.7554/eLife.48630
- ↑ Weis F, Menting JG, Margetts MB, Chan SJ, Xu Y, Tennagels N, Wohlfart P, Langer T, Muller CW, Dreyer MK, Lawrence MC. The signalling conformation of the insulin receptor ectodomain. Nat Commun. 2018 Oct 24;9(1):4420. doi: 10.1038/s41467-018-06826-6. PMID:30356040 doi:http://dx.doi.org/10.1038/s41467-018-06826-6
- ↑ Uchikawa E, Choi E, Shang G, Yu H, Bai XC. Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex. Elife. 2019 Aug 22;8. pii: 48630. doi: 10.7554/eLife.48630. PMID:31436533 doi:http://dx.doi.org/10.7554/eLife.48630
- ↑ Wilcox G. Insulin and insulin resistance. Clin Biochem Rev. 2005 May;26(2):19-39. PMID:16278749
- ↑ Riddle MC. Treatment of diabetes with insulin. From art to science. West J Med. 1983 Jun;138(6):838-46. PMID:6351440
Student Contributors
- Harrison Smith
- Alyssa Ritter