We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1606
From Proteopedia
(Difference between revisions)
| Line 23: | Line 23: | ||
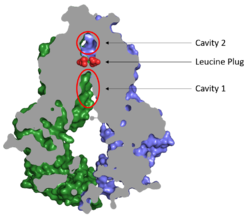
Substrates are transported through ABCG2 via two cavities separated by a leucine plug (Figure 2). <scene name='83/832932/Cavity_1_zoom_out/2'>Cavity 1</scene> acts as a multidrug binding pocket and is formed by helices at the interface of the monomers in the TMD. When ATP is not bound to the NBDs, Cavity 1 is <scene name='83/832932/Overall_structure_cavity_1hel/3'>open to the cytosol</scene> in order to recruit substrates for transport. Cavity 1 is <scene name='83/832932/Cavity_1_narrow_surface/4'>narrow</scene> and full of nonpolar, hydrophobic residues and, as a result, prefers nonpolar, hydrophobic substrates, particularly flat, polycyclic molecules. Substrates, such as estrone sulfate, <scene name='83/832932/Cavity_1_-_use2/4'>form hydrogen bonds and stacking interactions</scene> with residues from each subunit in Cavity 1.<ref name="Taylor"/> | Substrates are transported through ABCG2 via two cavities separated by a leucine plug (Figure 2). <scene name='83/832932/Cavity_1_zoom_out/2'>Cavity 1</scene> acts as a multidrug binding pocket and is formed by helices at the interface of the monomers in the TMD. When ATP is not bound to the NBDs, Cavity 1 is <scene name='83/832932/Overall_structure_cavity_1hel/3'>open to the cytosol</scene> in order to recruit substrates for transport. Cavity 1 is <scene name='83/832932/Cavity_1_narrow_surface/4'>narrow</scene> and full of nonpolar, hydrophobic residues and, as a result, prefers nonpolar, hydrophobic substrates, particularly flat, polycyclic molecules. Substrates, such as estrone sulfate, <scene name='83/832932/Cavity_1_-_use2/4'>form hydrogen bonds and stacking interactions</scene> with residues from each subunit in Cavity 1.<ref name="Taylor"/> | ||
| - | After substrates bind in Cavity 1, ATP binds each NBD leading to the transporter shifting from inward-facing to outward-facing. The outward-facing conformation results in the <scene name='83/832932/Atp_bound_cavity_2/3'>collapse of Cavity 1</scene> in the TMD which | + | After substrates bind in Cavity 1, ATP binds each NBD leading to the transporter shifting from inward-facing to outward-facing. The outward-facing conformation results in the <scene name='83/832932/Atp_bound_cavity_2/3'>collapse of Cavity 1</scene> in the TMD in which the cavity is no longer <scene name='83/832932/Overall_structure_cavity_1hel/3'>open to the cytosol</scene>. This collapse forces the substrate to move forward to Cavity 2 as there is no longer room in Cavity 1 to accommodate substrates.<ref name="Manolaridis"/> <scene name='83/832932/Atp_bound_use_cav_2/2'>Cavity 2</scene>, which is occluded when the protein in is the inward-facing conformation, is now open to the extracellular space and able to release the substrate. Cavity 2 contains a less hydrophobic environment and, as a result, substrates are released due to hydrophobic mismatch.<ref name="Taylor"/> <scene name='83/832932/Atp_bound_use_el_disulfides/3'>Disulfide bonds</scene> in the external loops near the exit of Cavity 2 also help promote substrate release.<ref name="Manolaridis"/> Once Cavity 2 is empty, the protein reverts to the inward-facing conformation via hydrolysis of ATP. |
Cavities 1 and 2 are separated by a <scene name='83/832932/Leucine_plug_open_con/7'>leucine plug</scene> which likely acts as a substrate check-point during transport; changes to either of these leucine residues have exhibited an increase in transport and a decrease in substrate specificity.<ref name="Manolaridis"/> After the substrate binds Cavity 1 and ATP molecules bind each NBD, the <scene name='83/832932/Atp_bound_cavsleu/5'>leucine plug opens</scene> to allow the substrate to enter Cavity 2. Once the substrate enters Cavity 2, the plug is able to reform and promote substrate release and conversion to the inward-facing conformation. | Cavities 1 and 2 are separated by a <scene name='83/832932/Leucine_plug_open_con/7'>leucine plug</scene> which likely acts as a substrate check-point during transport; changes to either of these leucine residues have exhibited an increase in transport and a decrease in substrate specificity.<ref name="Manolaridis"/> After the substrate binds Cavity 1 and ATP molecules bind each NBD, the <scene name='83/832932/Atp_bound_cavsleu/5'>leucine plug opens</scene> to allow the substrate to enter Cavity 2. Once the substrate enters Cavity 2, the plug is able to reform and promote substrate release and conversion to the inward-facing conformation. | ||
Revision as of 01:23, 17 April 2020
| This Sandbox is Reserved from Jan 13 through September 1, 2020 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1598 through Sandbox Reserved 1627. |
To get started:
More help: Help:Editing |
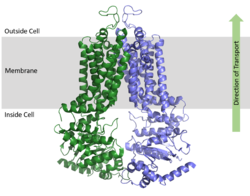
ABCG2 Multidrug Transporter
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Taylor NMI, Manolaridis I, Jackson SM, Kowal J, Stahlberg H, Locher KP. Structure of the human multidrug transporter ABCG2. Nature. 2017 Jun 22;546(7659):504-509. doi: 10.1038/nature22345. Epub 2017 May, 29. PMID:28554189 doi:http://dx.doi.org/10.1038/nature22345
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Manolaridis I, Jackson SM, Taylor NMI, Kowal J, Stahlberg H, Locher KP. Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states. Nature. 2018 Nov;563(7731):426-430. doi: 10.1038/s41586-018-0680-3. Epub 2018 Nov, 7. PMID:30405239 doi:http://dx.doi.org/10.1038/s41586-018-0680-3
- ↑ 3.0 3.1 Robey RW, Pluchino KM, Hall MD, Fojo AT, Bates SE, Gottesman MM. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat Rev Cancer. 2018 Jul;18(7):452-464. doi: 10.1038/s41568-018-0005-8. PMID:29643473 doi:http://dx.doi.org/10.1038/s41568-018-0005-8
- ↑ 4.0 4.1 4.2 4.3 Jackson SM, Manolaridis I, Kowal J, Zechner M, Taylor NMI, Bause M, Bauer S, Bartholomaeus R, Bernhardt G, Koenig B, Buschauer A, Stahlberg H, Altmann KH, Locher KP. Structural basis of small-molecule inhibition of human multidrug transporter ABCG2. Nat Struct Mol Biol. 2018 Apr;25(4):333-340. doi: 10.1038/s41594-018-0049-1. Epub, 2018 Apr 2. PMID:29610494 doi:http://dx.doi.org/10.1038/s41594-018-0049-1
Student Contributors
Julia Pomeroy