We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1619
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
===Background=== | ===Background=== | ||
| - | Gamma Secretase (GS) is a transmembrane [https://en.wikipedia.org/wiki/Aspartic_protease aspartatic protease]. It catalyzes peptide bond hydrolysis of type I integral membrane proteins such as Notch, amyloid precursor protein (APP), and various other substrates. It recognizes and catalyzes the reaction with its substrate using 3 residue segments. These 3 residue segments give rise to specific cleavage points for each substrate. APP is the primary substrate of focus, and it is cleaved to form amyloid-β peptides(Aβ). This product is important for various neural processes, and it is well known for its implications with [https://en.wikipedia.org/wiki/Alzheimer%27s_disease Alzheimer's disease (AD).] This has made GS a popular drug target, specifically using gamma secretase inhibitors. However, due to the nature of the enzyme having various neural functions, there are dangerous side effects when it is inhibited. | + | Gamma Secretase (GS) is a transmembrane [https://en.wikipedia.org/wiki/Aspartic_protease aspartatic protease]. It catalyzes peptide bond hydrolysis of type I integral membrane proteins such as Notch, amyloid precursor protein (APP), and various other substrates. It recognizes and catalyzes the reaction with its substrate using 3 residue segments. These 3 residue segments give rise to specific cleavage points for each substrate. APP is the primary substrate of focus, and it is cleaved to form amyloid-β peptides (Aβ). This product is important for various neural processes, and it is well known for its implications with [https://en.wikipedia.org/wiki/Alzheimer%27s_disease Alzheimer's disease (AD).] This has made GS a popular drug target, specifically using gamma secretase inhibitors. However, due to the nature of the enzyme having various neural functions, there are dangerous side effects when it is inhibited. |
===Overall Structure=== | ===Overall Structure=== | ||
| Line 13: | Line 13: | ||
<scene name='83/832945/Nct_subunit_shown/1'>NCT</scene> has a large extracellular domain and 1 TM. It is important to substrate recognition and binding. | <scene name='83/832945/Nct_subunit_shown/1'>NCT</scene> has a large extracellular domain and 1 TM. It is important to substrate recognition and binding. | ||
<scene name='83/832945/Ps1_subunit/1'>PS1</scene> serves as the active site of the protease and contains 9 TMs, each varying in length. The site of autocatalytic [https://en.wikipedia.org/wiki/Bond_cleavage cleavage] is located between <scene name='83/832945/Tm6_and_tm7_with_app/1'>TM6 and TM7</scene> in PS1. Major conformational changes take place in this subunit upon substrate binding. | <scene name='83/832945/Ps1_subunit/1'>PS1</scene> serves as the active site of the protease and contains 9 TMs, each varying in length. The site of autocatalytic [https://en.wikipedia.org/wiki/Bond_cleavage cleavage] is located between <scene name='83/832945/Tm6_and_tm7_with_app/1'>TM6 and TM7</scene> in PS1. Major conformational changes take place in this subunit upon substrate binding. | ||
| - | <scene name='83/832945/Aph-1_subunit/1'>APH-1</scene> serves as a scaffold for anchoring and supporting the flexible conformational changes of PS1 | + | <scene name='83/832945/Aph-1_subunit/1'>APH-1</scene> serves as a scaffold for anchoring and supporting the flexible conformational changes of PS1. |
Activation of the active site is dependent on the binding of <scene name='83/832945/Pen2_subunit/1'>PEN-2</scene>. PEN-2 is also important in maturation of the enzyme.<ref name="Yang">PMID:28628788</ref> | Activation of the active site is dependent on the binding of <scene name='83/832945/Pen2_subunit/1'>PEN-2</scene>. PEN-2 is also important in maturation of the enzyme.<ref name="Yang">PMID:28628788</ref> | ||
| Line 20: | Line 20: | ||
===Substrate Structure=== | ===Substrate Structure=== | ||
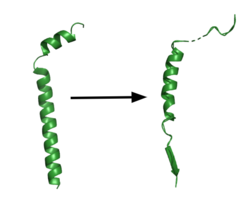
[[Image:App.png|250 px|right|thumb|'''Figure 1. APP fragment conformational change in gamma secretase.''' APP bound to gamma secretase undergoes a conformational change. The free state consists of 2 helices. The N-terminal helix unfolds into a coil and the C-terminal helix unwinds into a β-strand. This β-strand interacts with PS1 and is the site of cleavage by gamma secretase.]] | [[Image:App.png|250 px|right|thumb|'''Figure 1. APP fragment conformational change in gamma secretase.''' APP bound to gamma secretase undergoes a conformational change. The free state consists of 2 helices. The N-terminal helix unfolds into a coil and the C-terminal helix unwinds into a β-strand. This β-strand interacts with PS1 and is the site of cleavage by gamma secretase.]] | ||
| - | GS has multiple substrates, but the substrate of main concern is [https://en.wikipedia.org/wiki/Amyloid_precursor_protein/ APP]. APP is composed of an N-terminal loop and a | + | GS has multiple substrates, but the substrate of main concern is [https://en.wikipedia.org/wiki/Amyloid_precursor_protein/ APP]. APP is composed of an N-terminal loop and a transmembrane helix. It uses <scene name='83/832945/App_in_gs_general/1'>lateral diffusion</scene> as a mechanism of entry into the enzyme via the lid complex, and once in place, the TM helix is anchored by <scene name='83/832945/Hydrophobic_interactions/1'>van der Waals contacts in PS1</scene>. Upon binding to GS, the C-terminal extracellular helix unwinds. The N-terminal end of the TM helix unwinds into a β-strand (Fig. 1). In order to differentiate between different substrates like APP and notch, the β-strand is often the main point of identification for the enzyme. Substrate binding induces a structural change in GS, creating 2 β-strands that form a β-sheet with the substrate. This β-sheet is in close proximity with the active site, and guides the process of catalysis.<ref name="Zhou">PMID:30630874</ref>. |
===Lid Complex=== | ===Lid Complex=== | ||
| - | The <scene name='83/832945/Lidremake/1'>lid complex</scene> is the first point of entry and recognition for the substrate. This lid is located within the NCT subunit between Asn55 and Asn435. This lobe of NCT is divided into two separate subunits, being the large and small lobes. Phe287 from the large lobe acts as pivot between them. This <scene name='83/832945/Pivot/1'>Phe is further surrounded by Phe103, Leu171, Phe176, and Ile180</scene> of the small subunit, and these residues compose a greasy pocket | + | The <scene name='83/832945/Lidremake/1'>lid complex</scene> is the first point of entry and recognition for the substrate. This lid is located within the NCT subunit between Asn55 and Asn435. This lobe of NCT is divided into two separate subunits, being the large and small lobes. Phe287 from the large lobe acts as pivot between them. This <scene name='83/832945/Pivot/1'>Phe is further surrounded by Phe103, Leu171, Phe176, and Ile180</scene> of the small subunit, and these residues compose a greasy pocket that provides an environment for easy movement. The lid consists of 5 aromatic residues which are highly involved with stabilizing the closed conformation. In particular, this conformation is stabilized by <scene name='83/832945/Trp164scene/1'>Trp164, which interacts with Pro424, Phe448, and the aliphatic side chain of Gln420</scene>. Once the substrate binds and the lid is opened, a charged, hydrophilic pocket is revealed. This pocket contains <scene name='83/832945/Gluandtyr_remake/1'>Glu333 and Tyr337 surrounded by several charged residues</scene>, and is further involved with substrate binding and recognition once the lid is removed. However, this lid complex is relatively far away from the catalytic site of the enzyme in PS1. Once the substrate binds, the enzyme undergoes a conformational change to shorten this distance, and the rotation of the large lobe in relation to the small lobe reorients the substrate for cleavage by aligning the pocket in NCT to the active site in PS1. <ref name="Bai">PMID:26280335</ref> |
===Active Site=== | ===Active Site=== | ||
| - | The <scene name='83/832945/Asp_257_and_asp_385/12'>active site</scene> is located between TM6 and TM7 of the PS1 subunit, which is mainly hydrophilic and disordered. Each of these transmembrane helices has an aspartate residue, <scene name='83/832945/Asp_257_and_asp_385/10'>Asp257 and Asp385</scene>, which are located approximately 10.6 A˚ apart when inactive.<ref name="Bai">PMID:26280335</ref> The PAL sequence of <scene name='83/832945/Asp_257_and_asp_385/11'>Pro433, Ala434, and Leu435</scene> is in close proximity with the catalytic aspartates and is important to substrate recognition. GS becomes active upon substrate binding, when TM2 and TM6 each rotate about 15 degrees to more closely associate. Two β-strands are induced in PS1, creating an <scene name='83/832945/Beta_sheet_complex/1'>antiparallel β-sheet</scene> with the β-strand of APP. | + | The <scene name='83/832945/Asp_257_and_asp_385/12'>active site</scene> is located between TM6 and TM7 of the PS1 subunit, which is mainly hydrophilic and disordered. Each of these transmembrane helices has an aspartate residue, <scene name='83/832945/Asp_257_and_asp_385/10'>Asp257 and Asp385</scene>, which are located approximately 10.6 A˚ apart when inactive.<ref name="Bai">PMID:26280335</ref> The PAL sequence of <scene name='83/832945/Asp_257_and_asp_385/11'>Pro433, Ala434, and Leu435</scene> is in close proximity with the catalytic aspartates and is important to substrate recognition. GS becomes active upon substrate binding, when TM2 and TM6 each rotate about 15 degrees to more closely associate. Two β-strands are induced in PS1, creating an <scene name='83/832945/Beta_sheet_complex/1'>antiparallel β-sheet</scene> with the β-strand of APP.<ref name="Zhou" /> The β-strand of the substrate interacts via main chain H-bonds with the PAL sequence, stabilizing the active site. Asp257 and Asp385 hydrogen bond to each other and are located 6–7 Å away from the scissile peptide bond of the substrate, allowing catalysis to occur.<ref name="Yang" /> The cleavage site is between the helix and the N-terminal β-strand of APP. GS cleaves in 3 residue segments which is driven by the presence of three amino acid binding pockets in the active site. GS can cleave via different pathways, depending on its starting point, but the 2 most commonly used pathways produce Aβ48 and Aβ49.<ref name="Bolduc">PMID:27580372</ref>. Tripeptide cleavage starting between Thr719 and Leu720 results in Aβ48. Cleavage between Leu720 and Val721 yields Aβ49.<ref name="Zhou">PMID:30630874</ref> |
Revision as of 00:23, 21 April 2020
Gamma Secretase
| |||||||||||
References
- ↑ 1.0 1.1 Yang G, Zhou R, Shi Y. Cryo-EM structures of human gamma-secretase. Curr Opin Struct Biol. 2017 Oct;46:55-64. doi: 10.1016/j.sbi.2017.05.013. Epub, 2017 Jul 17. PMID:28628788 doi:http://dx.doi.org/10.1016/j.sbi.2017.05.013
- ↑ 2.0 2.1 2.2 2.3 Zhou R, Yang G, Guo X, Zhou Q, Lei J, Shi Y. Recognition of the amyloid precursor protein by human gamma-secretase. Science. 2019 Feb 15;363(6428). pii: science.aaw0930. doi:, 10.1126/science.aaw0930. Epub 2019 Jan 10. PMID:30630874 doi:http://dx.doi.org/10.1126/science.aaw0930
- ↑ 3.0 3.1 Bai XC, Yan C, Yang G, Lu P, Ma D, Sun L, Zhou R, Scheres SH, Shi Y. An atomic structure of human gamma-secretase. Nature. 2015 Aug 17. doi: 10.1038/nature14892. PMID:26280335 doi:http://dx.doi.org/10.1038/nature14892
- ↑ Bolduc DM, Montagna DR, Seghers MC, Wolfe MS, Selkoe DJ. The amyloid-beta forming tripeptide cleavage mechanism of gamma-secretase. Elife. 2016 Aug 31;5. doi: 10.7554/eLife.17578. PMID:27580372 doi:http://dx.doi.org/10.7554/eLife.17578
- ↑ Kumar D, Ganeshpurkar A, Kumar D, Modi G, Gupta SK, Singh SK. Secretase inhibitors for the treatment of Alzheimer's disease: Long road ahead. Eur J Med Chem. 2018 Mar 25;148:436-452. doi: 10.1016/j.ejmech.2018.02.035. Epub , 2018 Feb 15. PMID:29477076 doi:http://dx.doi.org/10.1016/j.ejmech.2018.02.035
Student Contributors
Layla Wisser
Daniel Mulawa