Overview
The mitochondrial calcium uniporter (MCU) complex is the main source of entry for calcium ions into the mitochondrial matrix from the intermembrane space. MCU channels exist in most eukaryotic life, but activity is regulated differently in each clade.[1] The precise identity of the MCU wasn't discovered until 2011 and was discovered using a combination of NMR spectroscopy, cryo-electron microscopy, and x-ray crystallography.[2] Identification of the structure was difficult because it has no apparent sequence similarity to other ion channels.[1] However, like other ion channels, it is incredibly selective and efficient. The MCU has the ability to only allow calcium ions into the mitochondrial matrix at a rate of 5,000,000 ions per second even though potassium ions are over 100,000 times more abundant in the intermembrane space.[1]
Under resting conditions, the calcium concentration in the mitochondria is about the same as in the cytoplasm, but when stimulated, it can increase calcium concentration 10-20-fold.[3] Mitochondria-associated ER membranes (MAMs) exist between mitochondria and the endoplasmic reticulum, the two largest cellular stores of calcium, to allow for efficient transport of calcium ions.[4] The transfer of electrons through respiratory complexes I-IV produces the energy to pump hydrogen ions into the intermembrane space (IMS) and create the proton electrochemical gradient potential.[3] This negative electrochemical potential is the driving force that moves positively charged calcium ions into the mitochondrial matrix.[3]
Regulation of the uptake and efflux of calcium is important to increase calcium levels enough to activate certain enzymes, but also avoid calcium overload and apoptosis.[4] Mitochondrial calcium increases ATP production by activating pyruvate dehydrogenase, α-ketoglutarate dehydrogenase, and isocitrate dehydrogenase in the Krebs cycle.[4] Therefore, deficiency of MCU leads to decrease of enzyme activity and of oxidative phosphorylation.
Structure
Mitochondrial Calcium Uniporter Complex
The mitochondrial calcium uniporter complex exists as a large complex (around 480 kDa in humans) made up of both pore-forming and regulatory subunits.[4] The mitochondrial uptake proteins (MICU1 and MICU2) are regulatory proteins in the MCU complex that exist in the IMS and contain EF hand domains for calcium binding to control transport through the channel of the MCU complex.[4] When calcium ion concentration in the IMS is low, MICU1 and 2 block the MCU to prevent uptake of calcium.[4] In the presence of high calcium concentrations, more calcium binds to these regulatory proteins and they undergo a conformational change to allow calcium ions through the MCU and into the matrix.[4] In fact, when calcium levels are below 500 nM, MICU1 can block movement of calcium by itself, calcium levels between 500 nM and 1,500 nM require both MICU1 and MICU2 to block ion entry, and any concentration over 1,500 nM is sufficient for calcium entry.[3] Another regulatory protein, MCUR1 is a cofactor in the assembly of the respiratory chain rather than an essential part of the uniporter.[3] Though the MCU is able to take up calcium independently, there are two other pore-forming subunits, the MCUb and the essential MCU regulator (EMRE).[4] MCUb is similar to MCU, but certain amino acids differ and make it an inhibitory subunit.[4] The EMRE is located in the IMS and connects MICU1 and MICU2 to the MCU.[3] It also contributes to regulation of calcium intake in the MCU.[4]
Mitochondrial Calcium Uniporter Structure
The mitochondrial calcium uniporter (MCU) denotes the channel protein itself, not to be confused with the MCU complex. The MCU was originally thought to be composed of a pentamer of five identical subunits, but it is now known to exist as a dimer of .[2] The protein as a whole is composed of a coiled-coil domain, a transmembrane domain, and a non-translated domain.[2] The hydrophobic is located in the inner mitochondrial membrane (IMM) and the hydrophilic coiled-coil domain exists in the mitochondrial matrix.[1] The transmembrane domain (TMD) consists of eight separate helices (TM1 and TM2 from each subunit) that are connected by mostly hydrophobic amino acids in the IMS and has four-fold symmetry.[1] packs tightly against from the neighboring subunit which conveys a sense of domain-swapping.[5] This section of the MCU can be roughly divided into a narrow outer leaflet portion with the selectivity filter and lined by the helices and a wide inner leaflet.[1] Past the transmembrane domain, the N-terminal domains of the helices extend into the matrix and form coiled-coils with a C-terminal helix.[1] These "legs" are separated from each other which allows enough space for calcium ions to diffuse out into the matrix.[1] Additionally, this domain is responsible for assembly of the MCU and post-translational modification.[5] Finally, each leg ends in a (NTD).[1] While the MCU can intake calcium without the NTD, it may have regulatory functions and the ability to bend transmembrane helices to constrict the pore.[1] [5]
Selectivity Filter

Fig. 1 Electronegativity of the MCU viewed from outside the mouth of the channel. The high concentration of negative charge (shown in red) attracts the positive character of calcium ions. Created using PyMOL.
The of the MCU is composed by many acidic amino acids near the narrow mouth of the channel which leads to high affinity for calcium (dissociation constant of less than 2nM).[1] The arrangement of the highly conserved motif in the TM2 helices form a ring in the pore to which calcium ions are attracted.[1] The structure in the animation is the MCU of Cyphellophora europaea so every amino acid named here specifically is that of C. europaea, but most of these residues are highly conserved across all species, though residue number may change. Though not part of the motif, is present at the mouth of the MCU and serves to congregate positively charged at the entrance of the channel.[1] The motif consists of at the N-terminal end, , , and .[1] pack against each other and are oriented towards the pore, but only serve to stabilize , not interact with calcium ions.[1][5] The X residues ( in this case) face away from the pore and are exposed to the membrane.[1] The negatively charged side chains of Asp225 and Glu228 point towards the pore and form rings of radius 2.5Å and 1Å, respectively.[1] It's a combination of these radii and charges that account for the selectivity of the MCU. For example, potassium has an ionic radius of 1.38Å which is much larger than the 1.00Å ionic radius of calcium and it therefore cannot fit into the negatively charged ring formed by Glu228.[1] Additionally, even though sodium ions have a similar ionic radius, the +2 charge on calcium is better matched to coordination with the glutamate residues.[1]
Movement of Calcium
Microscopy has revealed three sites in the MCU channel of roughly spherical density equally spaced 6Å apart.[1] Sites 1 and 2 lie within the selectivity filter so they can easily be assumed to contain calcium, but site 3 could be calcium or some other small molecule.[1] Site 1 is positioned in the ring formed by Asp225 residues and there is a distance of 4Å between the center of the site and each carboxylate group indicating presence of water.[1] The second site is positioned in the ring formed by Glu228 and there is a distance of 2.8Å between the carboxylate group of each residue and the middle of the site indicating absence of water.[1] It is hypothesized that one calcium ion coordinated with water in site 1 loses its water and moves to site 2 and a calcium ion moves from the IMS into site 1.[1] Meanwhile, a different calcium ion moves from site 2 to site 3 or the mitochondrial matrix.[1]
Mutations
There are a number of mutations that completely eliminate calcium uptake by the MCU. For example, mutation of W, D, E, or P of the WDXXEP motif altered the highly conserved selectivity filter and completely eliminated calcium uptake.[1][5] For example, even mutating Glu228 to an aspartate significantly changed the dimensions of the pore and inhibited uptake of calcium.[1] However, mutation of either X residue was not detrimental to calcium uptake.[1] Furthermore, mutation of a tyrosine residue directly below the selectivity filter substantially impaired calcium intake and proper protein folding.[5] The residue on TM1 that affected calcium uptake the most in human MCU was Trp317 (analogous to in C. europaea) which has a side chain constituting a primary contact point between TM1 and TM2.[5] Mutation of human MCU Phe326 (analogous to () in C. europaea) or Gly331 of the TM1-TM2 linker (() in C. europaea) affected the linker conformation and configuration of the pore entrance and impaired calcium intake.[5]
Regulation and Inhibition
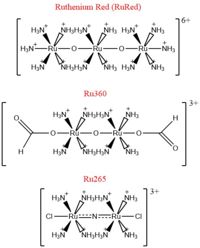
Fig. 2 Structures of the ruthenium-based inhibitors of the MCU. Created using ChemDraw Professional 16.0
The most well-known and commonly used inhibitor of the MCU is ruthenium red (RuRed).[2] RuRed is a trinuclear, oxo-bridged complex that effectively inhibits calcium uptake without affecting mitochondrial respiration or calcium efflux.[2] The disadvantage of ruthenium red is its challenging purification.[2] Interestingly, an impure version of RuRed, termed Ru360, was found to be the active component of RuRed and thus another good inhibitor of the MCU.[2] Ru360 is a binuclear, oxo-bridged complex with a similar structure to that of RuRed.[2] The only flaw with Ru360 was that it showed low cell permeability, so Ru265 was developed and had twice the cell permeability of Ru360.[2] Ru265 possesses two bridged Ru centers bridged by a nitride ligand.[2]
Recent experiments suggest that Ru360 inhibits calcium uptake through interactions with the WDXXEP motif.[2] However, not much is actually known about the method of inhibition. Mutations of Asp261 and Ser259 in human MCU (analogous to Asp225 and Ser223 in C. europaea) were shown to maintain calcium uptake into the matrix, but reduce the inhibitory effect of Ru360.[2] Curiously, the same Ser259 mutation did not affect inhibition of Ru265.[2] Additionally, a mutation in a cysteine residue in the NTD reduced the inhibitory effects of Ru265, but not Ru360.[2] So, while various inhibitors for the MCU are known, the mechanism of each is still largely unknown.
Medical Relevance
The MCU has a large role in disease because of its effect on apoptosis and cell signaling. The overload of the mitochondrial matrix with calcium leads to release of cytochrome c, overproduction of reactive oxygen species, mitochondrial swelling, and the opening of the mitochondrial permeability transition pore (mPTP) which all lead to apoptotic cell death.[2] This connection between mitochondrial calcium and apoptosis makes the MCU dysregulation a large contributor to cell death and disease. Calcium machinery in the mitochondria are targets for proto-oncogenes and tumor suppressors for this very reason.[3] Apoptosis can either be induced or repressed. Furthermore, external stimuli can activate receptors in the endoplasmic reticulum that release calcium and activate signal transductions.[4] Sequestration of calcium in the mitochondria is vital to shut down these activations, so any impact in movement of calcium ions can cause a wide variety of diseases.[4]
Neurodegenerative Disorders
Disruption in calcium homeostasis leads to a wide range of neurodegenerative disorders. The MCU complex has been identified to play a large role in neuromuscular disease because of a loss or mutation of MICU1.[2] This causes myopathy, learning difficulties, and progressive movement disorders which can be lethal.[2] In Alzheimer's disease, the buildup of amyloid-β plaques in the brain leads to increased calcium uptake in neurons and cell death.[2] Similarly, in early onset Parkinson's disease, degradation of MICU1 by the protein ligase Parkin leads to increased mitochondrial calcium uptake, overload, and death.[2] Finally, disrupted glutamate homeostasis in astrocytes and neurons leads to calcium overload and cell death via excitotoxicity in Amyotrophic Lateral Sclerosis (ALS).[2]
Diabetes
Calcium homeostasis dysregulation has also been proven to be instrumental in obesity, insulin resistance, and type-II diabetes.[4] The intracellular calcium concentrations in primary adipocytes from obese human subjects was found to be elevated.[4] Any inhbition of downstream calcium signaling could decrease movement of the GLUT4 glucose transporter and glucose uptake.[4] Additionally, ablation of MCU in β-cells in the pancreas demonstrated a decrease in cellular ATP concentration following glucose stimulation which resulted in decreased glucose-stimulated insulin secretion.[4] Furthermore, MAMs have been shown to cause glucose intolerance and mitochondrial dysfunction in primary hepatocytes in mice.[4] Subsequent reinforcement of these MAMs has been shown to increase insulin sensitivity and glucose homeostasis.[4]


