User:Alexis Walker/Sandbox 1
From Proteopedia
| Line 2: | Line 2: | ||
2V83 Recombination Activating Gene 2 | 2V83 Recombination Activating Gene 2 | ||
== Function == | == Function == | ||
| - | Recombination activating gene (RAG) 2 is a key protein in the rearrangement and recombination of genes that encode immunoglobulins and T-cell receptors. Both RAG1 and RAG2 form a V(D)J recombinase complex that is essential in maturing B- and T-cells, both of which are critical for adaptive immunity (Sadofsky). RAG cleaves dsDNA between the antigen receptor and the recombination signal sequence. These segments are then rejoined in a variable fashion that allows for the diversity within an organism's immune system. The recombination allows for the T- and B-cells to mature and ultimately act in their specified manner. More specifically RAG2 has a plant homeodomain (PHD) that recognizes a methylated lysine on histone H3. This domain is crucial to the function of RAG. | + | Recombination activating gene (RAG) 2 is a key nuclear protein in the rearrangement and recombination of genes that encode immunoglobulins and T-cell receptors. Both RAG1 and RAG2 form a V(D)J recombinase complex that is essential in maturing B- and T-cells, both of which are critical for adaptive immunity (Sadofsky). RAG cleaves dsDNA between the antigen receptor and the recombination signal sequence. These segments are then rejoined in a variable fashion that allows for the diversity within an organism's immune system. The recombination allows for the T- and B-cells to mature and ultimately act in their specified manner. More specifically RAG2 has a plant homeodomain (PHD) that recognizes a methylated lysine on histone H3. This domain is crucial to the function of RAG. |
== Disease == | == Disease == | ||
Five specific residues were implicated in patients with severe combined immunodeficiency (SCID). These residues are involved in histone recognition. (Ramón-Maiques) One mechanism is in the disruption of the overall protein structure results in T(-)B(-) SCID. This autosomal recessive disease is caused by an inability to form fully mature T- and B-cells, thus rendering them ineffective within the patient. Natural Killer cells are still present within the patient and the thalamus is typically hypoplastic. The condition can usually be treated with a bone marrow transplant. (Fischer) | Five specific residues were implicated in patients with severe combined immunodeficiency (SCID). These residues are involved in histone recognition. (Ramón-Maiques) One mechanism is in the disruption of the overall protein structure results in T(-)B(-) SCID. This autosomal recessive disease is caused by an inability to form fully mature T- and B-cells, thus rendering them ineffective within the patient. Natural Killer cells are still present within the patient and the thalamus is typically hypoplastic. The condition can usually be treated with a bone marrow transplant. (Fischer) | ||
| - | Failure to bind to H3K4me-3 Omenn Syndrome is a special type of SCID that is usually classified by a failure to thrive and elevated serum IgE levels. The autosomal recessive disease renders patients with low immunoglobulin levels and no B-cells. There are levels of T-cells in the blood but they are functionally impaired. (Aleman) Hypomorphic mutations of RAG contribute to Omenn Syndrome | + | Failure to bind to H3K4me-3 Omenn Syndrome is a special type of SCID that is usually classified by a failure to thrive and elevated serum IgE levels. The autosomal recessive disease renders patients with low immunoglobulin levels and no B-cells. There are levels of T-cells in the blood but they are functionally impaired. (Aleman) Hypomorphic mutations of RAG contribute to Omenn Syndrome. Studies have found that a specific W453R mutation alters the readout of H3K4me3 recognition (Matthews2), which is a direct molecular cause of Omenn Syndrome. |
Interestingly, infants born with the same RAG mutations can develop either SCID or Omenn Syndrome indicating that there may be an environmental component that contributes to disease development. | Interestingly, infants born with the same RAG mutations can develop either SCID or Omenn Syndrome indicating that there may be an environmental component that contributes to disease development. | ||
==Structure== | ==Structure== | ||
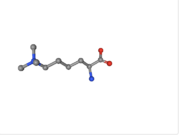
| - | 2V83 structure is composed of two total entities and five total chains. Within the VDJ Recombination Activating Protein 2 entity, there are a total of <scene name='84/842946/Vdj_component/1'>three chains</scene>. The second entity is the histone H3 entity. This is the region of the protein that binds to H3. There are a total of <scene name='84/842946/Chains_d_and_e/1'>two histone chains</scene>. There are a total of two N-trimethyllysines. | + | 2V83 structure is composed of two total entities and five total chains with a sequence length of 527(source). Within the VDJ Recombination Activating Protein 2 entity, there are a total of <scene name='84/842946/Vdj_component/1'>three chains</scene>. The second entity is the histone H3 entity. This is the region of the protein that binds to H3. There are a total of <scene name='84/842946/Chains_d_and_e/1'>two histone chains</scene>. There are a total of two N-trimethyllysines. |
[[Image:M3L.png | thumb | N-trimethyllysine]] | [[Image:M3L.png | thumb | N-trimethyllysine]] | ||
== Structural highlights == | == Structural highlights == | ||
RAG2 is bound to five total zinc ions which help to stabilize the DNA during recombination, <scene name='84/842946/Binding_site/1'>two of which</scene> (at residues 1487 and 1486) are involved in the DNA binding site. RAG2 contains a PHD near its C-terminus that specifically recognizes a methylated lysine 4 on histone H3. One unique component of the RAG2-PHD complex is that an peptide N-terminal to the RAG2-PHD can bind to the substrate-binding site. The ability of protein segments down the line to bind to an earlier section demonstrates the potential autoregulatory nature of this peptide(Ramón-Maiques). The peptide is also modified to yield enhanced binding to the H3K4me3. Normal interaction involves a carboxylate with an arginine 2 (R2). The RAG2-PHD replaces this carboxylate with a tyrosine (T445) which gives an interaction that is enhanced by the dimethylation of the R2 rather than inhibited. | RAG2 is bound to five total zinc ions which help to stabilize the DNA during recombination, <scene name='84/842946/Binding_site/1'>two of which</scene> (at residues 1487 and 1486) are involved in the DNA binding site. RAG2 contains a PHD near its C-terminus that specifically recognizes a methylated lysine 4 on histone H3. One unique component of the RAG2-PHD complex is that an peptide N-terminal to the RAG2-PHD can bind to the substrate-binding site. The ability of protein segments down the line to bind to an earlier section demonstrates the potential autoregulatory nature of this peptide(Ramón-Maiques). The peptide is also modified to yield enhanced binding to the H3K4me3. Normal interaction involves a carboxylate with an arginine 2 (R2). The RAG2-PHD replaces this carboxylate with a tyrosine (T445) which gives an interaction that is enhanced by the dimethylation of the R2 rather than inhibited. | ||
| + | |||
| + | ==Mechanism of Action== | ||
| + | RAG2 contains three residues that are primarily responsible for recognition of the tri-methyllysine present on histone H3. These residues form what is called the aromatic channel and are in positions, 415, 443, and 453 (Matthews2). The tyrosine, methionine and tryptophan residues surround the trimelthyllysine, identifying it for the reaction. Upon recognition the DNA is cleaved in a two-step process. First a nick is introduced at the 5' end of the recognized signal sequence. The complementary strand is then broken yielding a hairpin structure at the coding end (McBlane). Studies have shown that this mechanism can be done by purified forms of RAG1 and RAG2 only. | ||
== Evolution == | == Evolution == | ||
Revision as of 16:53, 28 April 2020
|
2V83 Recombination Activating Gene 2
Contents |
Function
Recombination activating gene (RAG) 2 is a key nuclear protein in the rearrangement and recombination of genes that encode immunoglobulins and T-cell receptors. Both RAG1 and RAG2 form a V(D)J recombinase complex that is essential in maturing B- and T-cells, both of which are critical for adaptive immunity (Sadofsky). RAG cleaves dsDNA between the antigen receptor and the recombination signal sequence. These segments are then rejoined in a variable fashion that allows for the diversity within an organism's immune system. The recombination allows for the T- and B-cells to mature and ultimately act in their specified manner. More specifically RAG2 has a plant homeodomain (PHD) that recognizes a methylated lysine on histone H3. This domain is crucial to the function of RAG.
Disease
Five specific residues were implicated in patients with severe combined immunodeficiency (SCID). These residues are involved in histone recognition. (Ramón-Maiques) One mechanism is in the disruption of the overall protein structure results in T(-)B(-) SCID. This autosomal recessive disease is caused by an inability to form fully mature T- and B-cells, thus rendering them ineffective within the patient. Natural Killer cells are still present within the patient and the thalamus is typically hypoplastic. The condition can usually be treated with a bone marrow transplant. (Fischer) Failure to bind to H3K4me-3 Omenn Syndrome is a special type of SCID that is usually classified by a failure to thrive and elevated serum IgE levels. The autosomal recessive disease renders patients with low immunoglobulin levels and no B-cells. There are levels of T-cells in the blood but they are functionally impaired. (Aleman) Hypomorphic mutations of RAG contribute to Omenn Syndrome. Studies have found that a specific W453R mutation alters the readout of H3K4me3 recognition (Matthews2), which is a direct molecular cause of Omenn Syndrome. Interestingly, infants born with the same RAG mutations can develop either SCID or Omenn Syndrome indicating that there may be an environmental component that contributes to disease development.
Structure
2V83 structure is composed of two total entities and five total chains with a sequence length of 527(source). Within the VDJ Recombination Activating Protein 2 entity, there are a total of . The second entity is the histone H3 entity. This is the region of the protein that binds to H3. There are a total of . There are a total of two N-trimethyllysines.
Structural highlights
RAG2 is bound to five total zinc ions which help to stabilize the DNA during recombination, (at residues 1487 and 1486) are involved in the DNA binding site. RAG2 contains a PHD near its C-terminus that specifically recognizes a methylated lysine 4 on histone H3. One unique component of the RAG2-PHD complex is that an peptide N-terminal to the RAG2-PHD can bind to the substrate-binding site. The ability of protein segments down the line to bind to an earlier section demonstrates the potential autoregulatory nature of this peptide(Ramón-Maiques). The peptide is also modified to yield enhanced binding to the H3K4me3. Normal interaction involves a carboxylate with an arginine 2 (R2). The RAG2-PHD replaces this carboxylate with a tyrosine (T445) which gives an interaction that is enhanced by the dimethylation of the R2 rather than inhibited.
Mechanism of Action
RAG2 contains three residues that are primarily responsible for recognition of the tri-methyllysine present on histone H3. These residues form what is called the aromatic channel and are in positions, 415, 443, and 453 (Matthews2). The tyrosine, methionine and tryptophan residues surround the trimelthyllysine, identifying it for the reaction. Upon recognition the DNA is cleaved in a two-step process. First a nick is introduced at the 5' end of the recognized signal sequence. The complementary strand is then broken yielding a hairpin structure at the coding end (McBlane). Studies have shown that this mechanism can be done by purified forms of RAG1 and RAG2 only.
Evolution
205 homologs were found via ConSurf-DB. Within chain A there is one predominant region with high levels of conservation. This region surrounds the N-trimethllysine. The same levels of conserved residues apply to chain B as well. Throughout multiple species the PHD-zinc finger is conserved, indicating that it is crucial to the function of RAG2. Due to its function, RAG2 is likely conserved across multiple species.
Links to 3-D Structures
Pre-reaction complex with DNA RAG 1/2 Core structure RAG2-PHD 2V83