User:Sumit Kamat/Sandbox Reserved 901
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
== Overview == | == Overview == | ||
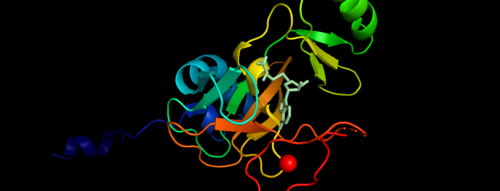
| - | Histone-lysine N-methyltransferase 2A (KMT2A) known as | + | Histone-lysine N-methyltransferase 2A (KMT2A) known as mixed-lineage leukemia 1 (MLL-1) is an enzyme that in humans is encoded by the KMT2A gene. KMT2A gene is a histone methyltransferase which are histone modifying enzymes which catalyze the transfer of methyl groups to lysine and arginine residues of histone proteins. The KMT2A gene is a positive global regulator of gene transcription and comprises of transactivation domain 9aaTAD which is involved in the epigenetic maintenance of transcriptional memory <ref>PMID 1720549</ref>. The KMT2 family can mono-, di- and trimethylates histone H3K4. This family of enzymes is found within a macromolecular complex known as the COMPASS family and are highly conserved from yeast to human <ref>PMC3711870</ref>. |
| Line 16: | Line 16: | ||
]] | ]] | ||
| + | == Structural Analysis == | ||
| + | |||
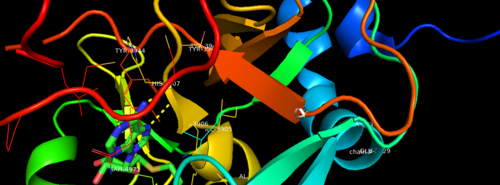
| + | MLL-1 is a 431-kDa protein to be a structural and functional homolog of the Drosophila trithorax (TRX) protein. Two domains are highly conserved between MLL and TRX consist of a carboxy-terminal SET (Su(var)3-9, enhancer-of-zeste, and trithorax) domain and internal plant homeodomain (PHD) fingers. Both domains are found in many chromatin-associated transcriptional regulators and are thought to function either directly in chromatin modification or as protein-protein interaction surfaces for the recruitment of chromatin-modifying machinery <ref> https://doi.org/10.1182/blood-2002-04-1015 </ref>. | ||
| Line 22: | Line 25: | ||
== Relevance == | == Relevance == | ||
| - | + | ||
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
Revision as of 17:50, 28 April 2020
Histone-lysine N-methyltransferase 2A KMT2A
| |||||||||||