This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Adenomatous polyposis coli
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
== The overall structure of APC == | == The overall structure of APC == | ||
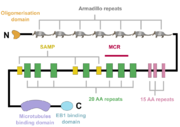
| - | The APC protein, its primary sequence encompassing 2843 aminoacids<ref name="uniprot">https://www.uniprot.org/uniprot/P25054</ref> | + | [[Image:APC.png|thumb|A schematic of the APC protein domain structure. MCR, mutation cluster region; SAMP, Axin-binding motif ]] |
| + | The APC protein, its primary sequence encompassing 2843 aminoacids<ref name="uniprot">https://www.uniprot.org/uniprot/P25054</ref>, consists of multiple domains, which enable it to interact with diverse partners. At the N-terminus, an oligomerisation domain is found, enabling the APC protein to oligomerise. It is followed by seven so called armadillo repeats, which form a groove for binding of a guanine nucleotide exchange factor Asef<ref name="Zhang2012">Zhang, Z. et al. (2012) ‘Structural basis for the recognition of Asef by adenomatous polyposis coli’, Cell Research. Nature Publishing Group, 22(2), pp. 372–386. doi: 10.1038/cr.2011.119.</ref>. The central part of APC contains three 15 aminoacid long repeats followed by seven 20 aminoacid long repeats<ref name="Zhang2017"/>. These motifs serve as binding sites for β-catenin<ref name="Hou2011">Hou, F. et al. (2011) ‘MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response.’, Cell. Elsevier, 146(3), pp. 448–61. doi: 10.1016/j.cell.2011.06.041.</ref>. In between the 20 aminoacid repeats, three SAMP regions are dispersed, enabling the interaction with Axin<ref name="Zhang2017"/>. At the C-terminus, a basic domain responsible for binding to microtubules as well as EB1 interaction domain are present<ref name="Su1995">Su, L. K. et al. (1995) ‘APC Binds to the Novel Protein EB’, Cancer Research, 55(14), pp. 2972–2977.</ref><ref name="Zhang2017"/>. | ||
Interestingly, majority of somatic mutations occurs in so called mutation cluster region (MCR) between codons 1286 and 1513 <ref name="Miyoshi1992">Miyoshi, Y. et al. (1992) Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene | Human Molecular Genetics | Oxford Academic, Human Molecular Genetics, Vol. 1, No. 4 229-233. Available at: https://academic.oup.com/hmg/article/1/4/229/730109 (Accessed: 22 April 2020).)</ref>. | Interestingly, majority of somatic mutations occurs in so called mutation cluster region (MCR) between codons 1286 and 1513 <ref name="Miyoshi1992">Miyoshi, Y. et al. (1992) Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene | Human Molecular Genetics | Oxford Academic, Human Molecular Genetics, Vol. 1, No. 4 229-233. Available at: https://academic.oup.com/hmg/article/1/4/229/730109 (Accessed: 22 April 2020).)</ref>. | ||
| - | + | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
== Disease == | == Disease == | ||
Revision as of 14:51, 29 April 2020
Adenomatous polyposis coli
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Zhang, L. and Shay, J. W. (2017) ‘Multiple Roles of APC and its Therapeutic Implications in Colorectal Cancer.’, Journal of the National Cancer Institute, 109(8). doi: 10.1093/jnci/djw332.
- ↑ https://www.proteinatlas.org/ENSG00000134982-APC/tissue
- ↑ Ficari, F. et al. (2000) ‘APC gene mutations and colorectal adenomatosis in familial adenomatous polyposis’, British Journal of Cancer. Churchill Livingstone, 82(2), pp. 348–353. doi: 10.1054/bjoc.1999.0925.
- ↑ Rowan, A. J. et al. (2000) ‘APC mutations in sporadic colorectal tumors: A mutational “hotspot” and interdependence of the “two hits”’, Proceedings of the National Academy of Sciences of the United States of America. National Academy of Sciences, 97(7), pp. 3352–3357. doi: 10.1073/pnas.97.7.3352.
- ↑ https://www.uniprot.org/uniprot/P25054
- ↑ Zhang, Z. et al. (2012) ‘Structural basis for the recognition of Asef by adenomatous polyposis coli’, Cell Research. Nature Publishing Group, 22(2), pp. 372–386. doi: 10.1038/cr.2011.119.
- ↑ Hou, F. et al. (2011) ‘MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response.’, Cell. Elsevier, 146(3), pp. 448–61. doi: 10.1016/j.cell.2011.06.041.
- ↑ Su, L. K. et al. (1995) ‘APC Binds to the Novel Protein EB’, Cancer Research, 55(14), pp. 2972–2977.
- ↑ Miyoshi, Y. et al. (1992) Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene | Human Molecular Genetics | Oxford Academic, Human Molecular Genetics, Vol. 1, No. 4 229-233. Available at: https://academic.oup.com/hmg/article/1/4/229/730109 (Accessed: 22 April 2020).)