This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Sumit Kamat/Sandbox Reserved 901
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
== Function == | == Function == | ||
| - | The KMT2A gene encodes a protein which is contains multiple conserved domains. KMT2A gene encodes a transcriptional coactivator that plays an important role in regulating gene expression during early development and hematopoiesis. Out of the many domains, SET domain is responsible for its histone H3 lysine 4 (H3K4) methyltransferase activity which mediates chromatin modifications associated with epigenetic transcriptional activation <ref> Otterness IG, Gans DJ. Nonsteroidal anti-inflammatory drugs: an analysis of the relationship between laboratory animal and clinical doses, including species scaling. Journal of Pharmaceutical Sciences. 1988 Sep;77(9):790-795. DOI: 10.1002/jps.2600770915. </ref>.The SET1 and MLL (KMT2) methyltransferases are conserved from yeast through humans. The Saccharomyces cerevisiae genome encodes a single H3K4 methyltransferase, Set1, whereas humans possess at least six homologs: SET1a, SET1b and MLL1-4. Unlike many SET domain enzymes, SET1 and MLL KMTs display very weak activity toward H3K4 and require additional subunits to attain maximal activity <ref> Nonsteroidal Anti-inflammatory Drugs: An Analysis of the Relationship between Laboratory Animal and Clinical Doses. Including Species Scaling | + | The KMT2A gene encodes a protein which is contains multiple conserved domains. KMT2A gene encodes a transcriptional coactivator that plays an important role in regulating gene expression during early development and hematopoiesis. Out of the many domains, SET domain is responsible for its histone H3 lysine 4 (H3K4) methyltransferase activity which mediates chromatin modifications associated with epigenetic transcriptional activation <ref> Otterness IG, Gans DJ. Nonsteroidal anti-inflammatory drugs: an analysis of the relationship between laboratory animal and clinical doses, including species scaling. Journal of Pharmaceutical Sciences. 1988 Sep;77(9):790-795. DOI: 10.1002/jps.2600770915. </ref>.The SET1 and MLL (KMT2) methyltransferases are conserved from yeast through humans. The Saccharomyces cerevisiae genome encodes a single H3K4 methyltransferase, Set1, whereas humans possess at least six homologs: SET1a, SET1b and MLL1-4. Unlike many SET domain enzymes, SET1 and MLL KMTs display very weak activity toward H3K4 and require additional subunits to attain maximal activity <ref> Nonsteroidal Anti-inflammatory Drugs: An Analysis of the Relationship between Laboratory Animal and Clinical Doses. Including Species Scaling Otterness, Ivan G. et al. Journal of Pharmaceutical Sciences, Volume 77, Issue 9, 790 - 795 </ref>.MLL-1 has a defined role in mammalian development, where it has been shown to be required for the regulation of important homeobox (Hox) genes and therefore is essential for early patterning in the embryo. In acute myeloid and lymphoid leukemia the MLL1 gene is frequently targeted in oncogenic gene translocations, leading to the expression of chimeric fusions between the amino-terminal 1400 residues of MLL1 and one of over 50 partner genes. MLL1 translocations are more prevalent in childhood leukemias and in therapy-related cases, where they are associated with a poor prognosis <ref> Guenther, M. G., Jenner, R. G., Chevalier, B., Nakamura, T., Croce, C. M., Canaani, E., & Young, R. A. (2005). Global and Hox-specific roles for the MLL1 methyltransferase. Proceedings of the National Academy of Sciences of the United States of America, 102(24), 8603–8608. https://doi.org/10.1073/pnas.0503072102 </ref>. |
| - | Otterness, Ivan G. et al. | + | |
| - | Journal of Pharmaceutical Sciences, Volume 77, Issue 9, 790 - 795 </ref>.MLL-1 has a defined role in mammalian development, where it has been shown to be required for the regulation of important homeobox (Hox) genes and therefore is essential for early patterning in the embryo. In acute myeloid and lymphoid leukemia the MLL1 gene is frequently targeted in oncogenic gene translocations, leading to the expression of chimeric fusions between the amino-terminal 1400 residues of MLL1 and one of over 50 partner genes. MLL1 translocations are more prevalent in childhood leukemias and in therapy-related cases, where they are associated with a poor prognosis <ref> Guenther, M. G., Jenner, R. G., Chevalier, B., Nakamura, T., Croce, C. M., Canaani, E., & Young, R. A. (2005). Global and Hox-specific roles for the MLL1 methyltransferase. Proceedings of the National Academy of Sciences of the United States of America, 102(24), 8603–8608. https://doi.org/10.1073/pnas.0503072102 </ref>. | + | |
| Line 28: | Line 26: | ||
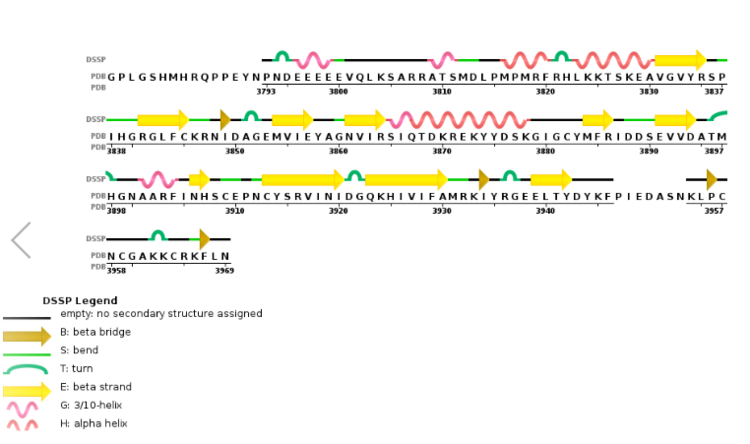
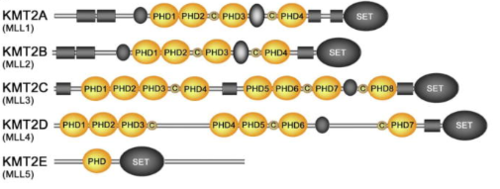
MLL-1 is a 431-kDa protein to be a structural and functional homolog of the Drosophila trithorax (TRX) protein. Two domains are highly conserved between MLL and TRX consist of a carboxy-terminal SET (Su(var)3-9, enhancer-of-zeste, and trithorax) domain and internal plant homeodomain (PHD) fingers. Both domains are found in many chromatin-associated transcriptional regulators and are thought to function either directly in chromatin modification or as protein-protein interaction surfaces for the recruitment of chromatin-modifying machinery <ref> Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties | MLL-1 is a 431-kDa protein to be a structural and functional homolog of the Drosophila trithorax (TRX) protein. Two domains are highly conserved between MLL and TRX consist of a carboxy-terminal SET (Su(var)3-9, enhancer-of-zeste, and trithorax) domain and internal plant homeodomain (PHD) fingers. Both domains are found in many chromatin-associated transcriptional regulators and are thought to function either directly in chromatin modification or as protein-protein interaction surfaces for the recruitment of chromatin-modifying machinery <ref> Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties | ||
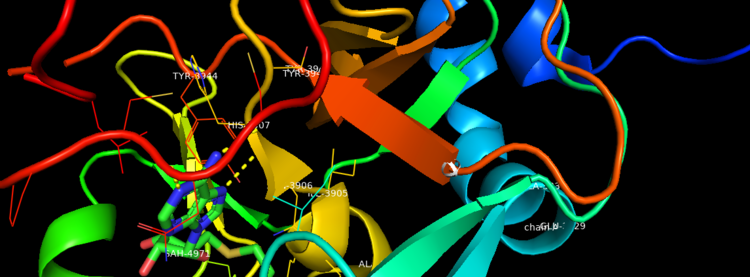
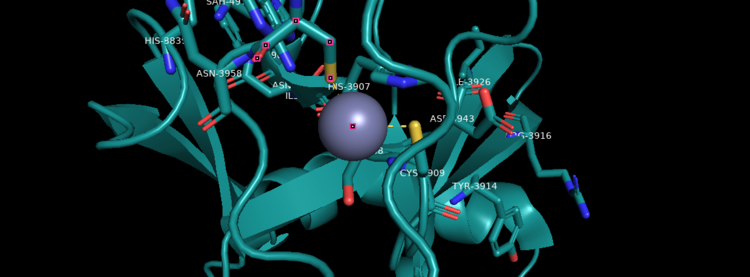
| - | Akihiko Yokoyama, Issay Kitabayashi, Paul M. Ayton, Michael L. Cleary, Misao Ohki https://doi.org/10.1182/blood-2002-04-1015 </ref>.There are no significant changes in the structure of the SET domain of the binary complex with the cofactor binding in the surface pocket. It has been determined at the essential active site residues like Phe3884, Tyr3942, Tyr3944, and Phe3946 and the main chain of the tetrapeptide, residues Cys3882 to Phe3885 have a similar arrangement to other SET domains earlier characterized. In the absence of substrate, the electron density for the side chain of Tyr3942 suggests two orientations, but upon substrate binding it becomes ordered and assumes the rotamer equivalent to other structures <ref> https://doi.org/10.1016/j.molcel.2008.12.029 </ref>. | + | Akihiko Yokoyama, Issay Kitabayashi, Paul M. Ayton, Michael L. Cleary, Misao Ohki https://doi.org/10.1182/blood-2002-04-1015 </ref>.There are no significant changes in the structure of the SET domain of the binary complex with the cofactor binding in the surface pocket. It has been determined at the essential active site residues like Phe3884, Tyr3942, Tyr3944, and Phe3946 and the main chain of the tetrapeptide, residues Cys3882 to Phe3885 have a similar arrangement to other SET domains earlier characterized. In the absence of substrate, the electron density for the side chain of Tyr3942 suggests two orientations, but upon substrate binding it becomes ordered and assumes the rotamer equivalent to other structures <ref> Structural Basis for the Requirement of Additional Factors for MLL1 SET Domain Activity and Recognition of Epigenetic Marks |
| + | Stacey M. Southall 2 Poon-Sheng Wong 2 Zain Odho S. Mark Roe Jon R. Wilson ArchiveDOI:https://doi.org/10.1016/j.molcel.2008.12.029 </ref>. | ||
Revision as of 23:11, 1 May 2020
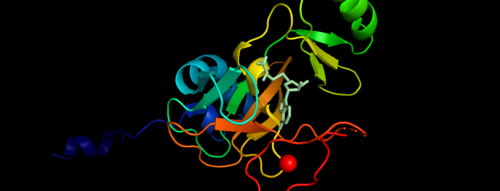
Histone-lysine N-methyltransferase 2A KMT2A
| |||||||||||