We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:R. Jeremy Johnson/Mitochondrial Calcium Uniporter
From Proteopedia
(Difference between revisions)
| Line 18: | Line 18: | ||
===Mitochondrial Calcium Uniporter Structure=== | ===Mitochondrial Calcium Uniporter Structure=== | ||
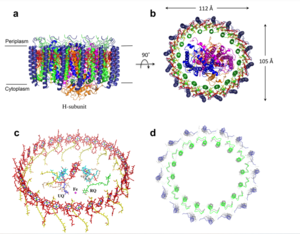
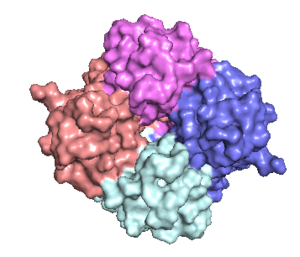
| - | The mitochondrial calcium uniporter (MCU) is the ion channel component. The MCU was originally thought to be composed of | + | The <scene name='83/832952/Starting_scene/5'>mitochondrial calcium uniporter (MCU)</scene> is the ion channel component (Figure 1). The MCU was originally thought to be composed of [https://en.wikipedia.org/wiki/Pentamer pentamer] of five identical subunits, but it is now known to exist as a [https://en.wikipedia.org/wiki/Dimer_(chemistry) dimer] of <scene name='83/832952/Dimer_of_dimers/5'>dimers</scene> (Figure 2).<ref name="Woods">PMID:31869674</ref> The <scene name='83/837230/Ntermsymmetry/1'>dimeric</scene> organization of the MCU is described as a <scene name='83/837230/Pyramid/2'>tetrameric truncated pyramid</scene>. The protein is composed of a <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene>, a <scene name='83/837230/Coiled_coil/3'>coiled coil domain</scene>, and a <scene name='83/837230/Nterm/2'>N-Terminal Domain</scene> (NTD) (Figure 1).<ref name="Woods"/> The hydrophobic <scene name='83/832952/New_ones/3'>transmembrane domain</scene> is located in the inner mitochondrial membrane ([https://en.wikipedia.org/wiki/Inner_mitochondrial_membrane IMM]) and the hydrophilic coiled-coil domain exists in the mitochondrial matrix.<ref name="Baradaran"/> |
===Transmembrane Domain=== | ===Transmembrane Domain=== | ||
| - | The <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene> is on the [https://en.wikipedia.org/wiki/Mitochondrion#Structure inner mitochondrial membrane] open to the inner membrane space. The small pore, highly specific for calcium binding is located in <scene name='83/837230/Tm2/1'>transmembrane 2</scene> (TM2) | + | The <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene> is on the [https://en.wikipedia.org/wiki/Mitochondrion#Structure inner mitochondrial membrane] open to the inner membrane space (Figure 1). The <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene> consists of eight separate helices (TM1 and TM2 from each subunit) that are connected by mostly hydrophobic amino acids in the IMS and has <scene name='83/832952/Starting_scene/5'>four-fold</scene> symmetry (Figure 4).<ref name="Baradaran"/> The small <scene name='83/832952/Selectivity_filter/3'>selectivity filter</scene> pore, highly specific for calcium binding is located in <scene name='83/837230/Tm2/1'>transmembrane 2</scene> (TM2) while <scene name='83/837230/Transmembrane_1/2'>transmembrane 1</scene> (TM1) surrounds the pore. <scene name='83/832952/Tm1/2'>TM1</scene> packs tightly against <scene name='83/832952/Tm2/2'>TM2</scene> from the neighboring subunit which conveys a sense of domain-swapping.<ref name="Fan">PMID:29995856</ref> The transmembrane domain of MCU can be roughly divided into a narrow outer leaflet portion with a <scene name='83/832952/Selectivity_filter/3'>selectivity filter</scene>, which is lined by the <scene name='83/832952/Tm2/2'>TM2</scene> helices and a wide inner leaflet.<ref name="Baradaran"/> |
===Coiled-coil domain=== | ===Coiled-coil domain=== | ||
| - | Past the transmembrane domain, the N-terminal domains of the <scene name='83/832952/Tm1/2'>TM1</scene> helices extend into the matrix and form coiled-coils with a C-terminal helix.<ref name="Baradaran"/> These "legs" are separated from each other which allows enough space for calcium ions to diffuse out into the matrix.<ref name="Baradaran"/> The <scene name='83/837230/Coiled_coil/3'>coiled coil</scene> is the first subsection of the soluble domain, which resides in the inner mitochondrial membrane. The coiled coil functions as the joints of the uniporter, providing flexibility to promote transport of | + | Past the transmembrane domain, the N-terminal domains of the <scene name='83/832952/Tm1/2'>TM1</scene> helices extend into the matrix and form coiled-coils with a C-terminal helix.<ref name="Baradaran"/> These "legs" are separated from each other which allows enough space for calcium ions to diffuse out into the matrix.<ref name="Baradaran"/> The <scene name='83/837230/Coiled_coil/3'>coiled coil</scene> domain is the first subsection of the soluble domain, which resides in the inner mitochondrial membrane. The coiled coil functions as the joints of the uniporter, providing flexibility to promote transport of Ca<sup>2+</sup>ions down their concentration gradient.<ref name="Fan" /> When Ca<sup>2+</sup> ions binds to the selectivity pore, the coiled-coil swings approximately 8° around its end near the <scene name='83/837230/Nterm/2'>NTD</scene>. This movement propagates to the top of the transmembrane domain, where the pore is located about 85 Å away. The largest displacement triggered by the movement of the coiled-coil is in the transmembrane domain, where the coil bends 20°, moving the transmembrane domain further apart. The junction between the transmembrane domain and the coiled coil's flexibility can be attributed to the disordered packing between subunits. Subunits A and C adopt different conformations than the B and D subunits, although they superimpose closely.<ref name="Fan" /> The coiled-coil domain is also responsible for assembly of the MCU and is [https://en.wikipedia.org/wiki/Post-translational_modification post-translationally modified].<ref name="Fan"/> |
[[Image:Nterm.png|300 px|right|thumb|Figure 2: Symmetry and organization of subunits from looking down into the uniporter from the inner mitochondrial membrane [https://en.wikipedia.org/wiki/Protein_Data_Bank PDB] [https://www.rcsb.org/structure/6DT0 6DT0]]] | [[Image:Nterm.png|300 px|right|thumb|Figure 2: Symmetry and organization of subunits from looking down into the uniporter from the inner mitochondrial membrane [https://en.wikipedia.org/wiki/Protein_Data_Bank PDB] [https://www.rcsb.org/structure/6DT0 6DT0]]] | ||
===N-terminal Domain=== | ===N-terminal Domain=== | ||
| - | Finally, each leg ends in a <scene name='83/832952/New_ones/4'>non-translated domain</scene> (NTD).<ref name="Baradaran"/> While the MCU can intake calcium without the NTD, | + | Finally, each leg ends in a <scene name='83/832952/New_ones/4'>non-translated domain</scene> (NTD).<ref name="Baradaran"/> While the MCU can intake calcium without the NTD, the NTD has regulatory functions, including bending <scene name='83/837230/Transmembrane_domain/3'>transmembrane helices</scene> to constrict the pore.<ref name="Baradaran"/><ref name="Fan"/> Reorganization in the NTD due to shifts in the The <scene name='83/837230/Coiled_coil/3'>coiled coil</scene> domain alters membrane packing, facilitating a rotamer switch between a pair of tyrosine residues controlling calcium flow through the pore. The soluble domain is wider than the transmembrane domain (Figure 1), allowing calcium ions to rehydrate and increasing the conductivity of ions through the uniporter into the mitochondrial matrix.<ref name="Fan" /> |
| - | + | ||
===Selectivity Filter=== | ===Selectivity Filter=== | ||
[[Image:Electronegativity_MCU_4.jpg|000 px|right|thumb|Figure3: Electronegativity of the MCU viewed from outside the channel. The high concentration of negative charge (shown in red) attracts the positive calcium ions. [https://en.wikipedia.org/wiki/Protein_Data_Bank PDB] [https://www.rcsb.org/structure/6DT0 6DT0]]] | [[Image:Electronegativity_MCU_4.jpg|000 px|right|thumb|Figure3: Electronegativity of the MCU viewed from outside the channel. The high concentration of negative charge (shown in red) attracts the positive calcium ions. [https://en.wikipedia.org/wiki/Protein_Data_Bank PDB] [https://www.rcsb.org/structure/6DT0 6DT0]]] | ||
| - | The <scene name='83/832952/Selectivity_filter/3'>selectivity filter</scene> of the MCU is composed by many acidic amino acids near the narrow mouth of the channel which leads to high affinity for calcium ([https://en.wikipedia.org/wiki/Dissociation_constant dissociation constant] of less than 2nM).<ref name="Baradaran"/> Negatively charged aspartates <scene name='83/832952/New_ones/2'>(Asp221)</scene> at the mouth of the MCU congregate positively charged <scene name='83/832952/Calcium/4'>calcium ions</scene> at the entrance of the channel.<ref name="Baradaran"/> A highly conserved <scene name='83/832952/Dxxe_motif/7'>WDXXEP</scene> [https://en.wikipedia.org/wiki/Sequence_motif motif] in the TM2 helices form the selectivity pore which selects for calcium transport over other similar ions.<ref name="Baradaran"/> | + | The <scene name='83/832952/Selectivity_filter/3'>selectivity filter</scene> of the MCU is composed by many acidic amino acids near the narrow mouth of the channel which leads to high affinity for calcium ([https://en.wikipedia.org/wiki/Dissociation_constant dissociation constant] of less than 2nM) (Figure 3).<ref name="Baradaran"/> Negatively charged aspartates <scene name='83/832952/New_ones/2'>(Asp221)</scene> at the mouth of the MCU congregate positively charged <scene name='83/832952/Calcium/4'>calcium ions</scene> at the entrance of the channel.<ref name="Baradaran"/> A highly conserved <scene name='83/832952/Dxxe_motif/7'>WDXXEP</scene> [https://en.wikipedia.org/wiki/Sequence_motif motif] in the TM2 helices form the selectivity pore which selects for calcium transport over other similar ions.<ref name="Baradaran"/> |
| - | The <scene name='83/832952/Dxxe_motif/7'>WDXXEP</scene> motif consists of <scene name='83/832952/Tryptophan/2'>Trp224</scene> at the N-terminal end, <scene name='83/832952/Selectivity_filter_asp/2'>Asp225</scene>, <scene name='83/832952/Selectivity_filter_glu/3'>Glu228</scene>, and <scene name='83/832952/New_ones/5'>Pro229</scene>.<ref name="Baradaran"/> The negatively charged side chains of Asp225 and <scene name='83/832933/Glu_358/4'>Glu228</scene> point towards the pore.<ref name="Baradaran"/> The <scene name='83/832933/Diameter/2'>diameter</scene> of the carboxyl ring is about 4Å, allowing only a dehydrated Ca<sup>2+</sup> ion to bind. The combination of these radii and high negative charge account for the selectivity of the MCU. For example, potassium has an [https://en.wikipedia.org/wiki/Ionic_radius ionic radius] of 1.38Å which is much larger than the 1.00Å ionic radius of calcium and potassium cannot fit into the negatively charged ring formed by <scene name='83/832952/Selectivity_filter_glu/3'>Glu228</scene>.<ref name="Baradaran"/> Additionally, even though sodium ions have a similar ionic radius, the +2 charge on calcium is better matched to coordination with the glutamate residues.<ref name="Baradaran"/> | + | The <scene name='83/832952/Dxxe_motif/7'>WDXXEP</scene> motif consists of <scene name='83/832952/Tryptophan/2'>Trp224</scene> at the N-terminal end, <scene name='83/832952/Selectivity_filter_asp/2'>Asp225</scene>, <scene name='83/832952/Selectivity_filter_glu/3'>Glu228</scene>, and <scene name='83/832952/New_ones/5'>Pro229</scene>.<ref name="Baradaran"/> The negatively charged side chains of Asp225 and <scene name='83/832933/Glu_358/4'>Glu228</scene> point towards the pore.<ref name="Baradaran"/> The <scene name='83/832933/Diameter/2'>diameter</scene> of the carboxyl ring is about 4Å, allowing only a dehydrated Ca<sup>2+</sup> ion to bind. The combination of these radii and high negative charge (Figure 3) account for the selectivity of the MCU. For example, potassium has an [https://en.wikipedia.org/wiki/Ionic_radius ionic radius] of 1.38Å which is much larger than the 1.00Å ionic radius of calcium and potassium cannot fit into the negatively charged ring formed by <scene name='83/832952/Selectivity_filter_glu/3'>Glu228</scene>.<ref name="Baradaran"/> Additionally, even though sodium ions have a similar ionic radius, the +2 charge on calcium is better matched to coordination with the glutamate residues.<ref name="Baradaran"/> |
scene name='83/832952/Tryptophan_proline/2'>Trp224 and Pro229</scene> pack against each other and are oriented towards the pore, but only serve to stabilize <scene name='83/832952/Selectivity_filter_glu/4'>Asp225 and Glu228</scene> and not interact with calcium ions.<ref name="Baradaran"/><ref name="Fan"/> Trp224 stabilizes the carbonyl side chains through <scene name='83/832933/H_bond_trp354_glu358/3'>hydrogen bonding</scene> and anion pi interactions. These Trp residues also form stacking interactions with Pro229, which orientate the Glu carboxyl side chains towards the middle of the pore to interact with Ca<sup>2+</sup> ions.<ref name=”Yoo”>PMID:29954988</ref> Approximately one helical turn below the glutamate ring of the selectivity filter, there is a tyrosine ring coming a 12Å wide pore allowing high conductivity. <ref name="Fan" /> The wider opening allows calcium to rehydrate once they pass the selectivity pore. | scene name='83/832952/Tryptophan_proline/2'>Trp224 and Pro229</scene> pack against each other and are oriented towards the pore, but only serve to stabilize <scene name='83/832952/Selectivity_filter_glu/4'>Asp225 and Glu228</scene> and not interact with calcium ions.<ref name="Baradaran"/><ref name="Fan"/> Trp224 stabilizes the carbonyl side chains through <scene name='83/832933/H_bond_trp354_glu358/3'>hydrogen bonding</scene> and anion pi interactions. These Trp residues also form stacking interactions with Pro229, which orientate the Glu carboxyl side chains towards the middle of the pore to interact with Ca<sup>2+</sup> ions.<ref name=”Yoo”>PMID:29954988</ref> Approximately one helical turn below the glutamate ring of the selectivity filter, there is a tyrosine ring coming a 12Å wide pore allowing high conductivity. <ref name="Fan" /> The wider opening allows calcium to rehydrate once they pass the selectivity pore. | ||
| Line 49: | Line 48: | ||
==Regulation and Inhibition== | ==Regulation and Inhibition== | ||
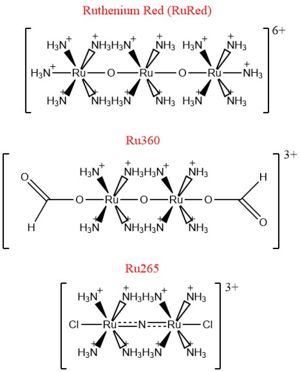
| - | [[Image:Ruthenium_Inhibitors.jpg|300 px|right|thumb| | + | [[Image:Ruthenium_Inhibitors.jpg|300 px|right|thumb|Figure 4: Structures of the ruthenium-based inhibitors of the MCU. Created using ChemDraw Professional 16.0]] |
The most well-known and commonly used inhibitor of the MCU is [https://en.wikipedia.org/wiki/Ruthenium_red ruthenium red] (RuRed).<ref name="Woods"/> RuRed is a trinuclear, oxo-bridged complex that effectively inhibits calcium uptake without affecting mitochondrial respiration or calcium efflux.<ref name="Woods"/> The disadvantage of ruthenium red is its challenging purification.<ref name="Woods"/> Interestingly, an impure version of RuRed, termed [https://en.wikipedia.org/wiki/Ru360 Ru360], was found to be the active component of RuRed and thus another good inhibitor of the MCU.<ref name="Woods"/> Ru360 is a binuclear, oxo-bridged complex with a similar structure to that of RuRed.<ref name="Woods"/> The only flaw with Ru360 was that it showed low cell permeability, so Ru265 was developed and had twice the cell permeability of Ru360.<ref name="Woods"/> Ru265 possesses two bridged Ru centers bridged by a nitride ligand.<ref name="Woods"/> | The most well-known and commonly used inhibitor of the MCU is [https://en.wikipedia.org/wiki/Ruthenium_red ruthenium red] (RuRed).<ref name="Woods"/> RuRed is a trinuclear, oxo-bridged complex that effectively inhibits calcium uptake without affecting mitochondrial respiration or calcium efflux.<ref name="Woods"/> The disadvantage of ruthenium red is its challenging purification.<ref name="Woods"/> Interestingly, an impure version of RuRed, termed [https://en.wikipedia.org/wiki/Ru360 Ru360], was found to be the active component of RuRed and thus another good inhibitor of the MCU.<ref name="Woods"/> Ru360 is a binuclear, oxo-bridged complex with a similar structure to that of RuRed.<ref name="Woods"/> The only flaw with Ru360 was that it showed low cell permeability, so Ru265 was developed and had twice the cell permeability of Ru360.<ref name="Woods"/> Ru265 possesses two bridged Ru centers bridged by a nitride ligand.<ref name="Woods"/> | ||
Revision as of 19:36, 8 May 2020
Mitochondrial Calcium Uniporter (MCU) Complex
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 1.21 1.22 1.23 1.24 1.25 1.26 1.27 Baradaran R, Wang C, Siliciano AF, Long SB. Cryo-EM structures of fungal and metazoan mitochondrial calcium uniporters. Nature. 2018 Jul 11. pii: 10.1038/s41586-018-0331-8. doi:, 10.1038/s41586-018-0331-8. PMID:29995857 doi:http://dx.doi.org/10.1038/s41586-018-0331-8
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 2.16 2.17 2.18 Woods JJ, Wilson JJ. Inhibitors of the mitochondrial calcium uniporter for the treatment of disease. Curr Opin Chem Biol. 2019 Dec 20;55:9-18. doi: 10.1016/j.cbpa.2019.11.006. PMID:31869674 doi:http://dx.doi.org/10.1016/j.cbpa.2019.11.006
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Giorgi C, Marchi S, Pinton P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat Rev Mol Cell Biol. 2018 Nov;19(11):713-730. doi: 10.1038/s41580-018-0052-8. PMID:30143745 doi:http://dx.doi.org/10.1038/s41580-018-0052-8
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 4.12 4.13 4.14 4.15 4.16 4.17 Wang CH, Wei YH. Role of mitochondrial dysfunction and dysregulation of Ca(2+) homeostasis in the pathophysiology of insulin resistance and type 2 diabetes. J Biomed Sci. 2017 Sep 7;24(1):70. doi: 10.1186/s12929-017-0375-3. PMID:28882140 doi:http://dx.doi.org/10.1186/s12929-017-0375-3
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 5.12 Fan C, Fan M, Orlando BJ, Fastman NM, Zhang J, Xu Y, Chambers MG, Xu X, Perry K, Liao M, Feng L. X-ray and cryo-EM structures of the mitochondrial calcium uniporter. Nature. 2018 Jul 11. pii: 10.1038/s41586-018-0330-9. doi:, 10.1038/s41586-018-0330-9. PMID:29995856 doi:http://dx.doi.org/10.1038/s41586-018-0330-9
- ↑ Yoo J, Wu M, Yin Y, Herzik MA Jr, Lander GC, Lee SY. Cryo-EM structure of a mitochondrial calcium uniporter. Science. 2018 Jun 28. pii: science.aar4056. doi: 10.1126/science.aar4056. PMID:29954988 doi:http://dx.doi.org/10.1126/science.aar4056
Student Contributors
Ryan Heumann
Lizzy Ratz
Holly Rowe
Madi Summers
Rieser Wells